Inferring differentially expressed pathways using kernel maximum mean discrepancy-based test
- PMID: 27294256
- PMCID: PMC4905616
- DOI: 10.1186/s12859-016-1046-1
Inferring differentially expressed pathways using kernel maximum mean discrepancy-based test
Abstract
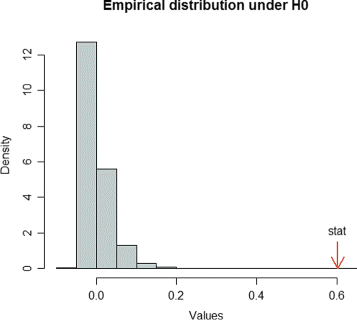
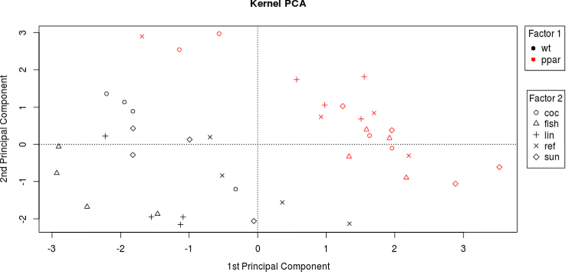
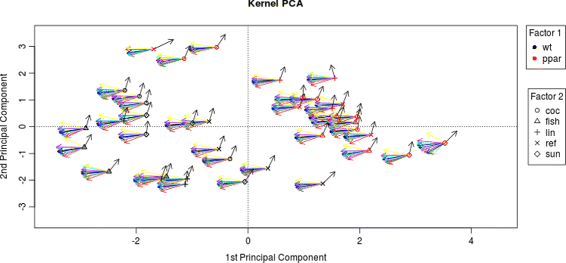
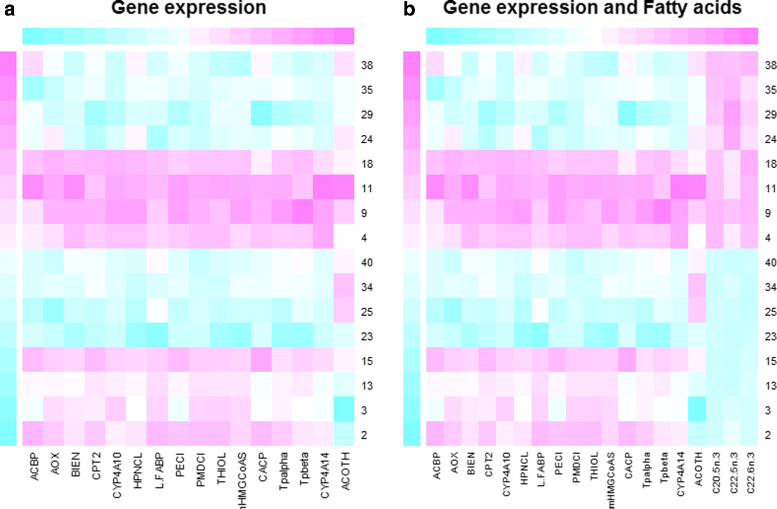
Background: Pathway expression is multivariate in nature. Thus, from a statistical perspective, to detect differentially expressed pathways between two conditions, methods for inferring differences between mean vectors need to be applied. Maximum mean discrepancy (MMD) is a statistical test to determine whether two samples are from the same distribution, its implementation being greatly simplified using the kernel method.
Results: An MMD-based test successfully detected the differential expression between two conditions, specifically the expression of a set of genes involved in certain fatty acid metabolic pathways. Furthermore, we exploited the ability of the kernel method to integrate data and successfully added hepatic fatty acid levels to the test procedure.
Conclusion: MMD is a non-parametric test that acquires several advantages when combined with the kernelization of data: 1) the number of variables can be greater than the sample size; 2) omics data can be integrated; 3) it can be applied not only to vectors, but to strings, sequences and other common structured data types arising in molecular biology.
Keywords: Kernel maximum mean test; Kernel-based methods; Omics data integration.
Figures

References
-
- Daemen A, Gevaert O, De Moor B. Integration of clinical and microarray data with kernel methods. In: Engineering in Medicine and Biology Society, 2007. EMBS 2007. 29th Annual International Conference of the IEEE: 2007. p. 5411–415. doi:10.1109/IEMBS.2007.4353566. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

