Integration of multi-omics data for prediction of phenotypic traits using random forest
- PMID: 27295212
- PMCID: PMC4905610
- DOI: 10.1186/s12859-016-1043-4
Integration of multi-omics data for prediction of phenotypic traits using random forest
Abstract
Background: In order to find genetic and metabolic pathways related to phenotypic traits of interest, we analyzed gene expression data, metabolite data obtained with GC-MS and LC-MS, proteomics data and a selected set of tuber quality phenotypic data from a diploid segregating mapping population of potato. In this study we present an approach to integrate these ~ omics data sets for the purpose of predicting phenotypic traits. This gives us networks of relatively small sets of interrelated ~ omics variables that can predict, with higher accuracy, a quality trait of interest.
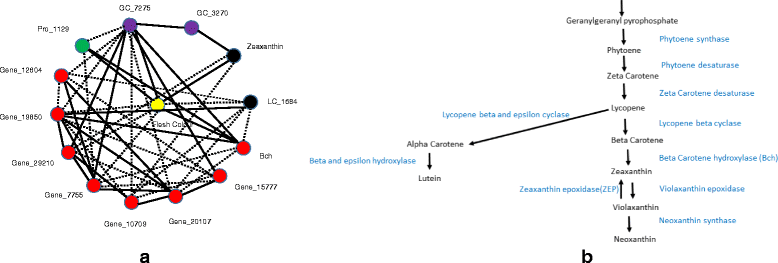
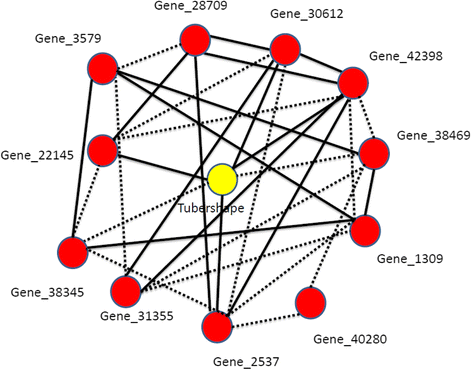
Results: We used Random Forest regression for integrating multiple ~ omics data for prediction of four quality traits of potato: tuber flesh colour, DSC onset, tuber shape and enzymatic discoloration. For tuber flesh colour beta-carotene hydroxylase and zeaxanthin epoxidase were ranked first and forty-fourth respectively both of which have previously been associated with flesh colour in potato tubers. Combining all the significant genes, LC-peaks, GC-peaks and proteins, the variation explained was 75 %, only slightly more than what gene expression or LC-MS data explain by themselves which indicates that there are correlations among the variables across data sets. For tuber shape regressed on the gene expression, LC-MS, GC-MS and proteomics data sets separately, only gene expression data was found to explain significant variation. For DSC onset, we found 12 significant gene expression, 5 metabolite levels (GC) and 2 proteins that are associated with the trait. Using those 19 significant variables, the variation explained was 45 %. Expression QTL (eQTL) analyses showed many associations with genomic regions in chromosome 2 with also the highest explained variation compared to other chromosomes. Transcriptomics and metabolomics analysis on enzymatic discoloration after 5 min resulted in 420 significant genes and 8 significant LC metabolites, among which two were putatively identified as caffeoylquinic acid methyl ester and tyrosine.
Conclusions: In this study, we made a strategy for selecting and integrating multiple ~ omics data using random forest method and selected representative individual peaks for networks based on eQTL, mQTL or pQTL information. Network analysis was done to interpret how a particular trait is associated with gene expression, metabolite and protein data.
Keywords: Data integration; Genetical genomics; Networks; Random forest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

