A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles
- PMID: 27295279
- PMCID: PMC4905634
- DOI: 10.1371/journal.ppat.1005672
A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles
Abstract
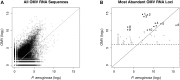
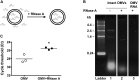
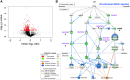
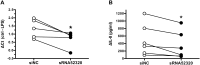
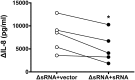
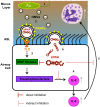
Bacterial outer membrane vesicle (OMV)-mediated delivery of proteins to host cells is an important mechanism of host-pathogen communication. Emerging evidence suggests that OMVs contain differentially packaged short RNAs (sRNAs) with the potential to target host mRNA function and/or stability. In this study, we used RNA-Seq to characterize differentially packaged sRNAs in Pseudomonas aeruginosa OMVs, and to show transfer of OMV sRNAs to human airway cells. We selected one sRNA for further study based on its stable secondary structure and predicted mRNA targets. Our candidate sRNA (sRNA52320), a fragment of a P. aeruginosa methionine tRNA, was abundant in OMVs and reduced LPS-induced as well as OMV-induced IL-8 secretion by cultured primary human airway epithelial cells. We also showed that sRNA52320 attenuated OMV-induced KC cytokine secretion and neutrophil infiltration in mouse lung. Collectively, these findings are consistent with the hypothesis that sRNA52320 in OMVs is a novel mechanism of host-pathogen interaction whereby P. aeruginosa reduces the host immune response.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
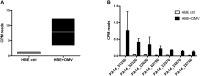

References
-
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013 [Internet]. 2013. Available: http://www.cdc.gov/drugresistance/threat-report-2013/index.html
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

