Cancer metabolism as a central driving force of glioma pathogenesis
- PMID: 27295313
- PMCID: PMC5488809
- DOI: 10.1007/s10014-016-0265-5
Cancer metabolism as a central driving force of glioma pathogenesis
Abstract
The recent identification of distinct genetic and epigenetic features in each glioma entity is leading to a multilayered, integrated diagnostic approach combining histologic features with molecular genetic information. Somatic mutations in isocitrate dehydrogenase (IDH) and receptor tyrosine kinase (RTK) pathways are key oncogenic events in diffuse gliomas, including lower grade (grade II and III) gliomas (LGG) and the highly lethal brain tumor glioblastoma (GBM), respectively, where they reprogram the epigenome, transcriptome, and metabolome to drive tumor growth. However, the mechanisms by which these genetic aberrations are translated into the aggressive nature of gliomas through metabolic reprogramming have just begun to be unraveled. The intricate interactions between the oncogenic signaling and cancer metabolism have also been recently demonstrated. Here, we describe a set of recent discoveries on cancer metabolism driven by IDH mutation and mutations in RTK pathways, highlighting the integration of genetic mutations, metabolic reprogramming, and epigenetic shifts, potentially providing new therapeutic opportunities.
Keywords: Genetic-metabolism interaction; Glioma; IDH; Metabolic reprogramming; Molecular genetics; RTK.
Figures
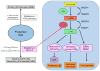
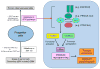
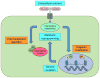
References
-
- Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. - PubMed
-
- Arita H, Narita Y, Yoshida A, Hashimoto N, Yoshimine T, Ichimura K. IDH1/2 mutation detection in gliomas. Brain Tumor Pathol. 2015;32:79–89. - PubMed
-
- Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B, Zhu S, Gu Y, Villa GR, Akhavan D, Nathanson D, Gini B, Mareninov S, Li R, Camacho CE, Kurdistani SK, Eskin A, Nelson SF, Yong WH, Cavenee WK, Cloughesy TF, Christofk HR, Black DL, Mischel PS. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17:1000–1008. - PMC - PubMed
-
- Bai H, Harmancı AS, Erson-Omay EZ, Li J, Coşkun S, Simon M, Krischek B, Özduman K, Omay SB, Sorensen EA, Turcan Ş, Bakırcığlu M, Carrión-Grant G, Murray PB, Clark VE, Ercan-Sencicek AG, Knight J, Sencar L, Altınok S, Kaulen LD, Gülez B, Timmer M, Schramm J, Mishra-Gorur K, Henegariu O, Moliterno J, Louvi A, Chan TA, Tannheimer SL, Pamir MN, Vortmeyer AO, Bilguvar K, Yasuno K, Günel M. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48:59–66. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

