RNA sequencing analysis to capture the transcriptome landscape during skin ulceration syndrome progression in sea cucumber Apostichopus japonicus
- PMID: 27296384
- PMCID: PMC4906609
- DOI: 10.1186/s12864-016-2810-3
RNA sequencing analysis to capture the transcriptome landscape during skin ulceration syndrome progression in sea cucumber Apostichopus japonicus
Abstract
Background: Sea cucumber Apostichopus japonicus is an important economic species in China, which is affected by various diseases; skin ulceration syndrome (SUS) is the most serious. In this study, we characterized the transcriptomes in A. japonicus challenged with Vibrio splendidus to elucidate the changes in gene expression throughout the three stages of SUS progression.
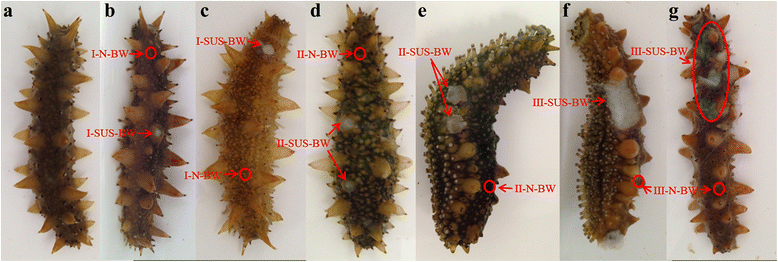
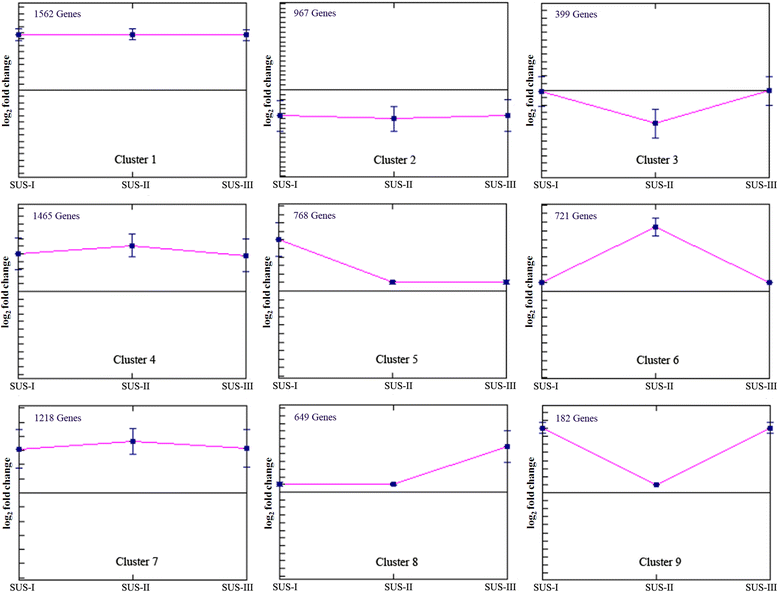
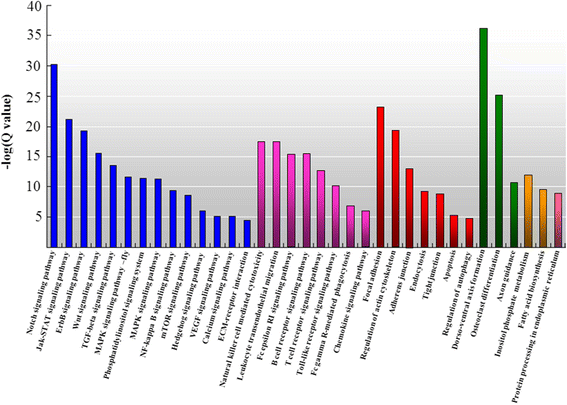
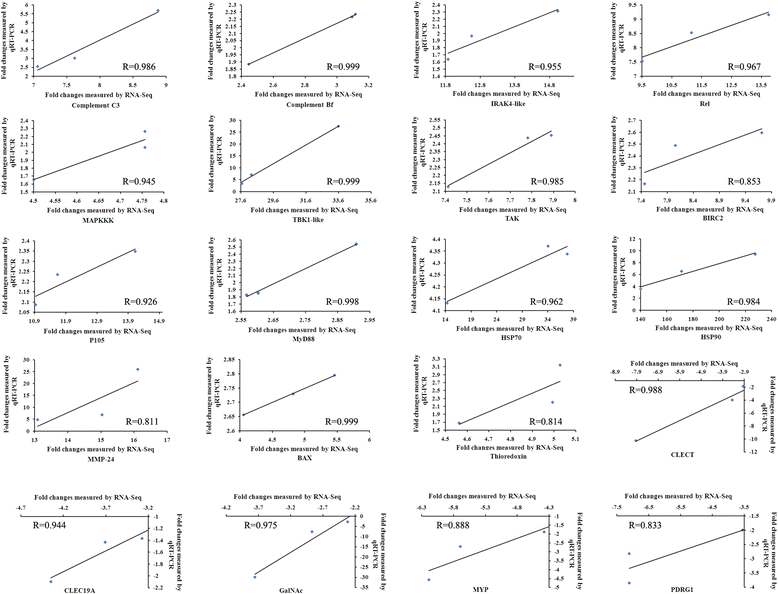
Results: RNA sequencing of 21 cDNA libraries from various tissues and developmental stages of SUS-affected A. japonicus yielded 553 million raw reads, of which 542 million high-quality reads were generated by deep-sequencing using the Illumina HiSeq™ 2000 platform. The reference transcriptome comprised a combination of the Illumina reads, 454 sequencing data and Sanger sequences obtained from the public database to generate 93,163 unigenes (average length, 1,052 bp; N50 = 1,575 bp); 33,860 were annotated. Transcriptome comparisons between healthy and SUS-affected A. japonicus revealed greater differences in gene expression profiles in the body walls (BW) than in the intestines (Int), respiratory trees (RT) and coelomocytes (C). Clustering of expression models revealed stable up-regulation as the main pattern occurring in the BW throughout the three stages of SUS progression. Significantly affected pathways were associated with signal transduction, immune system, cellular processes, development and metabolism. Ninety-two differentially expressed genes (DEGs) were divided into four functional categories: attachment/pathogen recognition (17), inflammatory reactions (38), oxidative stress response (7) and apoptosis (30). Using quantitative real-time PCR, twenty representative DEGs were selected to validate the sequencing results. The Pearson's correlation coefficient (R) of the 20 DEGs ranged from 0.811 to 0.999, which confirmed the consistency and accuracy between these two approaches.
Conclusions: Dynamic changes in global gene expression occur during SUS progression in A. japonicus. Elucidation of these changes is important in clarifying the molecular mechanisms associated with the development of SUS in sea cucumber.
Keywords: Dynamic expression profiles; RNA sequencing; Sea cucumber (Apostichopus japonicus); Skin ulceration syndrome.
Figures
Similar articles
-
Transcriptome sequencing of sea cucumber (Apostichopus japonicus) and the identification of gene-associated markers.Mol Ecol Resour. 2014 Jan;14(1):127-38. doi: 10.1111/1755-0998.12147. Epub 2013 Jul 16. Mol Ecol Resour. 2014. PMID: 23855518
-
Proteomic analysis reveals the important roles of alpha-5-collagen and ATP5β during skin ulceration syndrome progression of sea cucumber Apostichopus japonicus.J Proteomics. 2018 Mar 20;175:136-143. doi: 10.1016/j.jprot.2018.01.001. Epub 2018 Jan 8. J Proteomics. 2018. PMID: 29325989
-
Expression analysis of microRNAs related to the skin ulceration syndrome of sea cucumber Apostichopus japonicus.Fish Shellfish Immunol. 2016 Feb;49:205-12. doi: 10.1016/j.fsi.2015.12.036. Epub 2015 Dec 24. Fish Shellfish Immunol. 2016. PMID: 26723265
-
Transcriptome analysis provides insights into the molecular mechanisms responsible for evisceration behavior in the sea cucumber Apostichopus japonicus.Comp Biochem Physiol Part D Genomics Proteomics. 2019 Jun;30:143-157. doi: 10.1016/j.cbd.2019.02.008. Epub 2019 Feb 27. Comp Biochem Physiol Part D Genomics Proteomics. 2019. PMID: 30851504
-
Application of Computational Biology to Decode Brain Transcriptomes.Genomics Proteomics Bioinformatics. 2019 Aug;17(4):367-380. doi: 10.1016/j.gpb.2019.03.003. Epub 2019 Oct 23. Genomics Proteomics Bioinformatics. 2019. PMID: 31655213 Free PMC article. Review.
Cited by
-
Insights into high-pressure acclimation: comparative transcriptome analysis of sea cucumber Apostichopus japonicus at different hydrostatic pressure exposures.BMC Genomics. 2020 Jan 21;21(1):68. doi: 10.1186/s12864-020-6480-9. BMC Genomics. 2020. PMID: 31964339 Free PMC article.
-
Transcriptomic Responses of a Lightly Calcified Echinoderm to Experimental Seawater Acidification and Warming during Early Development.Biology (Basel). 2023 Dec 13;12(12):1520. doi: 10.3390/biology12121520. Biology (Basel). 2023. PMID: 38132346 Free PMC article.
-
Genome-Wide Identification of Circular RNAs Revealed the Dominant Intergenic Region Circularization Model in Apostichopus japonicus.Front Genet. 2019 Jul 2;10:603. doi: 10.3389/fgene.2019.00603. eCollection 2019. Front Genet. 2019. PMID: 31312211 Free PMC article.
-
Sea Cucumber Body Vesicular Syndrome Is Driven by the Pond Water Microbiome via an Altered Gut Microbiota.mSystems. 2022 Jun 28;7(3):e0135721. doi: 10.1128/msystems.01357-21. Epub 2022 Apr 14. mSystems. 2022. PMID: 35418244 Free PMC article.
-
Sea cucumbers: an emerging system in evo-devo.Evodevo. 2024 Feb 17;15(1):3. doi: 10.1186/s13227-023-00220-0. Evodevo. 2024. PMID: 38368336 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

