An assay for 26S proteasome activity based on fluorescence anisotropy measurements of dye-labeled protein substrates
- PMID: 27296635
- PMCID: PMC4976823
- DOI: 10.1016/j.ab.2016.05.026
An assay for 26S proteasome activity based on fluorescence anisotropy measurements of dye-labeled protein substrates
Abstract
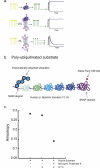
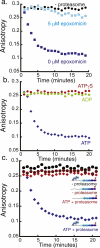
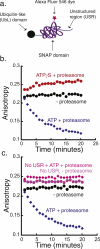
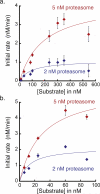
The 26S proteasome is the molecular machine at the center of the ubiquitin proteasome system and is responsible for adjusting the concentrations of many cellular proteins. It is a drug target in several human diseases, and assays for the characterization of modulators of its activity are valuable. The 26S proteasome consists of two components: a core particle, which contains the proteolytic sites, and regulatory caps, which contain substrate receptors and substrate processing enzymes, including six ATPases. Current high-throughput assays of proteasome activity use synthetic fluorogenic peptide substrates that report directly on the proteolytic activity of the proteasome, but not on the activities of the proteasome caps that are responsible for protein recognition and unfolding. Here, we describe a simple and robust assay for the activity of the entire 26S proteasome using fluorescence anisotropy to follow the degradation of fluorescently labeled protein substrates. We describe two implementations of the assay in a high-throughput format and show that it meets the expected requirement of ATP hydrolysis and the presence of a canonical degradation signal or degron in the target protein.
Keywords: 26S proteasome; Fluorescence anisotropy; High-throughput degradation assay; Protein degradation; Ubiquitin proteasome system (UPS).
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
References
-
- Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009;10:104–115. - PubMed
-
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

