Lack of FTSH4 Protease Affects Protein Carbonylation, Mitochondrial Morphology, and Phospholipid Content in Mitochondria of Arabidopsis: New Insights into a Complex Interplay
- PMID: 27297677
- PMCID: PMC4972270
- DOI: 10.1104/pp.16.00370
Lack of FTSH4 Protease Affects Protein Carbonylation, Mitochondrial Morphology, and Phospholipid Content in Mitochondria of Arabidopsis: New Insights into a Complex Interplay
Erratum in
-
CORRECTION: Vol. 171: 2516-2535, 2016.Plant Physiol. 2016 Oct;172(2):1352. doi: 10.1104/pp.16.01303. Plant Physiol. 2016. PMID: 27694396 Free PMC article. No abstract available.
Abstract
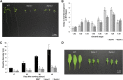
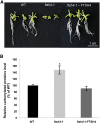
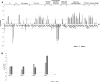
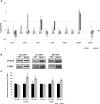
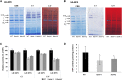
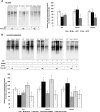
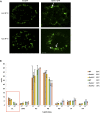
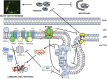
FTSH4 is one of the inner membrane-embedded ATP-dependent metalloproteases in mitochondria of Arabidopsis (Arabidopsis thaliana). In mutants impaired to express FTSH4, carbonylated proteins accumulated and leaf morphology was altered when grown under a short-day photoperiod, at 22°C, and a long-day photoperiod, at 30°C. To provide better insight into the function of FTSH4, we compared the mitochondrial proteomes and oxyproteomes of two ftsh4 mutants and wild-type plants grown under conditions inducing the phenotypic alterations. Numerous proteins from various submitochondrial compartments were observed to be carbonylated in the ftsh4 mutants, indicating a widespread oxidative stress. One of the reasons for the accumulation of carbonylated proteins in ftsh4 was the limited ATP-dependent proteolytic capacity of ftsh4 mitochondria, arising from insufficient ATP amount, probably as a result of an impaired oxidative phosphorylation (OXPHOS), especially complex V. In ftsh4, we further observed giant, spherical mitochondria coexisting among normal ones. Both effects, the increased number of abnormal mitochondria and the decreased stability/activity of the OXPHOS complexes, were probably caused by the lower amount of the mitochondrial membrane phospholipid cardiolipin. We postulate that the reduced cardiolipin content in ftsh4 mitochondria leads to perturbations within the OXPHOS complexes, generating more reactive oxygen species and less ATP, and to the deregulation of mitochondrial dynamics, causing in consequence the accumulation of oxidative damage.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures

References
-
- Anand R, Langer T, Baker MJ (2013) Proteolytic control of mitochondrial function and morphogenesis. Biochim Biophys Acta 1833: 195–204 - PubMed
-
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 - PMC - PubMed
-
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR (2011) Protein degradation: an alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498 - PubMed
-
- Augustin S, Nolden M, Müller S, Hardt O, Arnold I, Langer T (2005) Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J Biol Chem 280: 2691–2699 - PubMed
-
- Baker MJ, Mooga VP, Guiard B, Langer T, Ryan MT, Stojanovski D (2012) Impaired folding of the mitochondrial small TIM chaperones induces clearance by the i-AAA protease. J Mol Biol 424: 227–239 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

