Integrated mate-pair and RNA sequencing identifies novel, targetable gene fusions in peripheral T-cell lymphoma
- PMID: 27297792
- PMCID: PMC5009513
- DOI: 10.1182/blood-2016-03-707141
Integrated mate-pair and RNA sequencing identifies novel, targetable gene fusions in peripheral T-cell lymphoma
Abstract
Peripheral T-cell lymphomas (PTCLs) represent a heterogeneous group of T-cell malignancies that generally demonstrate aggressive clinical behavior, often are refractory to standard therapy, and remain significantly understudied. The most common World Health Organization subtype is PTCL, not otherwise specified (NOS), essentially a "wastebasket" category because of inadequate understanding to assign cases to a more specific diagnostic entity. Identification of novel fusion genes has contributed significantly to improving the classification, biologic understanding, and therapeutic targeting of PTCLs. Here, we integrated mate-pair DNA and RNA next-generation sequencing to identify chromosomal rearrangements encoding expressed fusion transcripts in PTCL, NOS. Two of 11 cases had novel fusions involving VAV1, encoding a truncated form of the VAV1 guanine nucleotide exchange factor important in T-cell receptor signaling. Fluorescence in situ hybridization studies identified VAV1 rearrangements in 10 of 148 PTCLs (7%). These were observed exclusively in PTCL, NOS (11%) and anaplastic large cell lymphoma (11%). In vitro, ectopic expression of a VAV1 fusion promoted cell growth and migration in a RAC1-dependent manner. This growth was inhibited by azathioprine, a clinically available RAC1 inhibitor. We also identified novel kinase gene fusions, ITK-FER and IKZF2-ERBB4, as candidate therapeutic targets that show similarities to known recurrent oncogenic ITK-SYK fusions and ERBB4 transcript variants in PTCLs, respectively. Additional novel and potentially clinically relevant fusions also were discovered. Together, these findings identify VAV1 fusions as recurrent and targetable events in PTCLs and highlight the potential for clinical sequencing to guide individualized therapy approaches for this group of aggressive malignancies.
© 2016 by The American Society of Hematology.
Figures
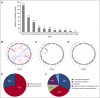

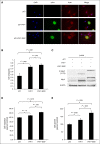

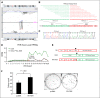
Comment in
-
Targetable gene fusions in T-cell lymphoma.Blood. 2016 Sep 1;128(9):1161-2. doi: 10.1182/blood-2016-07-723999. Blood. 2016. PMID: 27587864 No abstract available.
References
-
- Armitage JO. The aggressive peripheral T-cell lymphomas: 2012 update on diagnosis, risk stratification, and management. Am J Hematol. 2012;87(5):511–519. - PubMed
-
- Vose J, Armitage J, Weisenburger D International T-Cell Lymphoma Project. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26(25):4124–4130. - PubMed
-
- Abouyabis AN, Shenoy PJ, Lechowicz MJ, Flowers CR. Incidence and outcomes of the peripheral T-cell lymphoma subtypes in the United States. Leuk Lymphoma. 2008;49(11):2099–2107. - PubMed
-
- Pileri SA, et al. Peripheral T-cell lymphoma, not otherwise specified. In: Swerdlow S, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer; 2008. pp. 306–308.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

