Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma
- PMID: 27297799
- PMCID: PMC4906518
- DOI: 10.1038/srep27859
Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma
Abstract
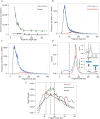
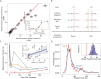
Circulating cell-free DNA (cfDNA) is emerging as a powerful monitoring tool in cancer, pregnancy and organ transplantation. Nucleosomal DNA, the predominant form of plasma cfDNA, can be adapted for sequencing via ligation of double-stranded DNA (dsDNA) adapters. dsDNA library preparations, however, are insensitive to ultrashort, degraded cfDNA. Drawing inspiration from advances in paleogenomics, we have applied a single-stranded DNA (ssDNA) library preparation method to sequencing of cfDNA in the plasma of lung transplant recipients (40 samples, six patients). We found that ssDNA library preparation yields a greater portion of sub-100 bp nuclear genomic cfDNA (p 10(-5), Mann-Whitney U Test), and an increased relative abundance of mitochondrial (10.7x, p 10(-5)) and microbial cfDNA (71.3x, p 10(-5)). The higher yield of microbial sequences from this method increases the sensitivity of cfDNA-based monitoring for infections following transplantation. We detail the fragmentation pattern of mitochondrial, nuclear genomic and microbial cfDNA over a broad fragment length range. We report the observation of donor-specific mitochondrial cfDNA in the circulation of lung transplant recipients. A ssDNA library preparation method provides a more informative window into understudied forms of cfDNA, including mitochondrial and microbial derived cfDNA and short nuclear genomic cfDNA, while retaining information provided by standard dsDNA library preparation methods.
Conflict of interest statement
Cornell University has applied for a patent relating to methods described in this study.
Figures


References
-
- Lo Y. D. et al. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet 351, 1329–1330 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

