AtDOF5.4/OBP4, a DOF Transcription Factor Gene that Negatively Regulates Cell Cycle Progression and Cell Expansion in Arabidopsis thaliana
- PMID: 27297966
- PMCID: PMC4906354
- DOI: 10.1038/srep27705
AtDOF5.4/OBP4, a DOF Transcription Factor Gene that Negatively Regulates Cell Cycle Progression and Cell Expansion in Arabidopsis thaliana
Abstract
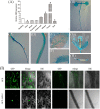
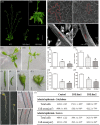
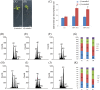
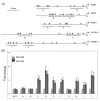
In contrast to animals, plant development involves continuous organ formation, which requires strict regulation of cell proliferation. The core cell cycle machinery is conserved across plants and animals, but plants have developed new mechanisms that precisely regulate cell proliferation in response to internal and external stimuli. Here, we report that the DOF transcription factor OBP4 negatively regulates cell proliferation and expansion. OBP4 is a nuclear protein. Constitutive and inducible overexpression of OBP4 reduced the cell size and number, resulting in dwarf plants. Inducible overexpression of OBP4 in Arabidopsis also promoted early endocycle onset and inhibited cell expansion, while inducible overexpression of OBP4 fused to the VP16 activation domain in Arabidopsis delayed endocycle onset and promoted plant growth. Furthermore, gene expression analysis showed that cell cycle regulators and cell wall expansion factors were largely down-regulated in the OBP4 overexpression lines. Short-term inducible analysis coupled with in vivo ChIP assays indicated that OBP4 targets the CyclinB1;1, CDKB1;1 and XTH genes. These results strongly suggest that OBP4 is a negative regulator of cell cycle progression and cell growth. These findings increase our understanding of the transcriptional regulation of the cell cycle in plants.
Figures



Similar articles
-
The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana.Plant J. 2008 Dec;56(5):779-92. doi: 10.1111/j.1365-313X.2008.03641.x. Epub 2008 Sep 4. Plant J. 2008. PMID: 18665917
-
ABA Suppresses Root Hair Growth via the OBP4 Transcriptional Regulator.Plant Physiol. 2017 Mar;173(3):1750-1762. doi: 10.1104/pp.16.01945. Epub 2017 Feb 6. Plant Physiol. 2017. PMID: 28167701 Free PMC article.
-
The transcription factor OBP4 controls root growth and promotes callus formation.New Phytol. 2017 Mar;213(4):1787-1801. doi: 10.1111/nph.14315. Epub 2016 Nov 17. New Phytol. 2017. PMID: 27859363
-
Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants.Plant Cell Physiol. 2004 Apr;45(4):386-91. doi: 10.1093/pcp/pch055. Plant Cell Physiol. 2004. PMID: 15111712 Review.
-
The plant cell cycle: an overview.Methods Mol Biol. 2005;296:31-50. doi: 10.1385/1-59259-857-9:031. Methods Mol Biol. 2005. PMID: 15576925 Review.
Cited by
-
Interspaced Repeat Sequences Confer the Regulatory Functions of AtXTH10, Important for Root Growth in Arabidopsis.Plants (Basel). 2019 May 16;8(5):130. doi: 10.3390/plants8050130. Plants (Basel). 2019. PMID: 31100875 Free PMC article.
-
Comparative transcriptomics identifies candidate genes involved in the evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae).BMC Plant Biol. 2022 Jul 14;22(1):340. doi: 10.1186/s12870-022-03631-8. BMC Plant Biol. 2022. PMID: 35836106 Free PMC article.
-
Genome-Wide Identification and Analysis of DOF Gene Family in Eugenia uniflora L. (Myrtaceae).Genes (Basel). 2022 Nov 28;13(12):2235. doi: 10.3390/genes13122235. Genes (Basel). 2022. PMID: 36553502 Free PMC article.
-
Identification of Two Auxin-Regulated Potassium Transporters Involved in Seed Maturation.Int J Mol Sci. 2018 Jul 22;19(7):2132. doi: 10.3390/ijms19072132. Int J Mol Sci. 2018. PMID: 30037141 Free PMC article.
-
DNA-Binding One Finger Transcription Factor PhDof28 Regulates Petal Size in Petunia.Int J Mol Sci. 2023 Jul 26;24(15):11999. doi: 10.3390/ijms241511999. Int J Mol Sci. 2023. PMID: 37569375 Free PMC article.
References
-
- Aggarwal P. et al.. The TCP4 transcription factor of Arabidopsis blocks cell division in yeast at G1–>S transition. Biochem Biophys Res Commun 410, 276–281 (2011). - PubMed
-
- Bernstein H. S. & Coughlin S. R. A mammalian homolog of fission yeast Cdc5 regulates G2 progression and mitotic entry. Journal of Biological Chemistry 273, 4666–4671 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

