Resolving the etiology of atopic disorders by using genetic analysis of racial ancestry
- PMID: 27297995
- PMCID: PMC5014679
- DOI: 10.1016/j.jaci.2016.02.045
Resolving the etiology of atopic disorders by using genetic analysis of racial ancestry
Abstract
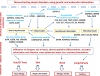
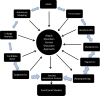
Atopic dermatitis (AD), food allergy, allergic rhinitis, and asthma are common atopic disorders of complex etiology. The frequently observed atopic march from early AD to asthma, allergic rhinitis, or both later in life and the extensive comorbidity of atopic disorders suggest common causal mechanisms in addition to distinct ones. Indeed, both disease-specific and shared genomic regions exist for atopic disorders. Their prevalence also varies among races; for example, AD and asthma have a higher prevalence in African Americans when compared with European Americans. Whether this disparity stems from true genetic or race-specific environmental risk factors or both is unknown. Thus far, the majority of the genetic studies on atopic diseases have used populations of European ancestry, limiting their generalizability. Large-cohort initiatives and new analytic methods, such as admixture mapping, are currently being used to address this knowledge gap. Here we discuss the unique and shared genetic risk factors for atopic disorders in the context of ancestry variations and the promise of high-throughput "-omics"-based systems biology approach in providing greater insight to deconstruct their genetic and nongenetic etiologies. Future research will also focus on deep phenotyping and genotyping of diverse racial ancestry, gene-environment, and gene-gene interactions.
Keywords: -omics; Atopic march; admixture mapping; allergic rhinitis; asthma; atopic dermatitis; food allergy; gene-environment interaction; phenotyping; racial ancestry.
Copyright © 2016 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Statement M.E.R. is a consultant for Genentech, Novartis, Receptos, and NKT Therapeutics and has an equity interest in NKT Therapeutics, Immune Pharmaceuticals, and Celsus Therapeutics, as well as royalty interest from Teva Pharmaceuticals and Cincinnati Children’s Medical Center–owned patents concerning eosinophilic esophagitis. The rest of the authors declare that they have no competing interests.
Figures









References
-
- Spergel JM, Paller AS. Atopic dermatitis and the atopic march. The Journal of allergy and clinical immunology. 2003 Dec;112(6 Suppl):S118–127. - PubMed
-
- Spergel JM. Atopic march: link to upper airways. Current opinion in allergy and clinical immunology. 2005 Feb;5(1):17–21. - PubMed
-
- Akdis CA. Allergy and hypersensitivity: mechanisms of allergic disease. Current opinion in immunology. 2006 Dec;18(6):718–726. - PubMed
-
- Litonjua AA, Celedon JC, Hausmann J, et al. Variation in total and specific IgE: effects of ethnicity and socioeconomic status. The Journal of allergy and clinical immunology. 2005 Apr;115(4):751–757. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

