S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation
- PMID: 27298336
- PMCID: PMC4911931
- DOI: 10.1101/gad.280834.116
S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation
Abstract
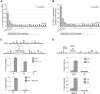
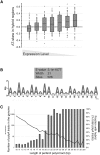
R loops form when transcripts hybridize to homologous DNA on chromosomes, yielding a DNA:RNA hybrid and a displaced DNA single strand. R loops impact the genome of many organisms, regulating chromosome stability, gene expression, and DNA repair. Understanding the parameters dictating R-loop formation in vivo has been hampered by the limited quantitative and spatial resolution of current genomic strategies for mapping R loops. We report a novel whole-genome method, S1-DRIP-seq (S1 nuclease DNA:RNA immunoprecipitation with deep sequencing), for mapping hybrid-prone regions in budding yeast Saccharomyces cerevisiae Using this methodology, we identified ∼800 hybrid-prone regions covering 8% of the genome. Given the pervasive transcription of the yeast genome, this result suggests that R-loop formation is dictated by characteristics of the DNA, RNA, and/or chromatin. We successfully identified two features highly predictive of hybrid formation: high transcription and long homopolymeric dA:dT tracts. These accounted for >60% of the hybrid regions found in the genome. We demonstrated that these two factors play a causal role in hybrid formation by genetic manipulation. Thus, the hybrid map generated by S1-DRIP-seq led to the identification of the first global genomic features causal for R-loop formation in yeast.
Keywords: DNA:RNA hybrids; R loops; RNase H; polyA tracts; transcription.
© 2016 Wahba et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ. 2000. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells 5: 789–802. - PubMed
-
- Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. 2013. Telomeric RNA–DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol 20: 1199–1205. - PubMed
-
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. 1986. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods 89: 123–130. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
