Activation of the molecular chaperone, sigma 1 receptor, preserves cone function in a murine model of inherited retinal degeneration
- PMID: 27298364
- PMCID: PMC4932934
- DOI: 10.1073/pnas.1521749113
Activation of the molecular chaperone, sigma 1 receptor, preserves cone function in a murine model of inherited retinal degeneration
Abstract
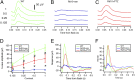
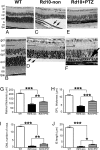
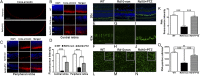
Retinal degenerative diseases are major causes of untreatable blindness, and novel approaches to treatment are being sought actively. Here we explored the activation of a unique protein, sigma 1 receptor (Sig1R), in the treatment of PRC loss because of its multifaceted role in cellular survival. We used Pde6β(rd10) (rd10) mice, which harbor a mutation in the rod-specific phosphodiesterase gene Pde6β and lose rod and cone photoreceptor cells (PRC) within the first 6 wk of life, as a model for severe retinal degeneration. Systemic administration of the high-affinity Sig1R ligand (+)-pentazocine [(+)-PTZ] to rd10 mice over several weeks led to the rescue of cone function as indicated by electroretinographic recordings using natural noise stimuli and preservation of cone cells upon spectral domain optical coherence tomography and retinal histological examination. The protective effect appears to result from the activation of Sig1R, because rd10/Sig1R(-/-) mice administered (+)-PTZ exhibited no cone preservation. (+)-PTZ treatment was associated with several beneficial cellular phenomena including attenuated reactive gliosis, reduced microglial activation, and decreased oxidative stress in mutant retinas. To our knowledge, this is the first report that activation of Sig1R attenuates inherited PRC loss. The findings may have far-reaching therapeutic implications for retinal neurodegenerative diseases.
Keywords: (+)-pentazocine; oxidative stress; photoreceptor; rd10 mouse; retinal neuroprotection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
The molecular chaperone sigma 1 receptor mediates rescue of retinal cone photoreceptor cells via modulation of NRF2.Free Radic Biol Med. 2019 Apr;134:604-616. doi: 10.1016/j.freeradbiomed.2019.02.001. Epub 2019 Feb 10. Free Radic Biol Med. 2019. PMID: 30743048 Free PMC article.
-
Optimal timing for activation of sigma 1 receptor in the Pde6brd10/J (rd10) mouse model of retinitis pigmentosa.Exp Eye Res. 2021 Jan;202:108397. doi: 10.1016/j.exer.2020.108397. Epub 2020 Dec 9. Exp Eye Res. 2021. PMID: 33310057 Free PMC article.
-
Activation of Sigma 1 Receptor Extends Survival of Cones and Improves Visual Acuity in a Murine Model of Retinitis Pigmentosa.Invest Ophthalmol Vis Sci. 2019 Oct 1;60(13):4397-4407. doi: 10.1167/iovs.19-27709. Invest Ophthalmol Vis Sci. 2019. PMID: 31639826 Free PMC article.
-
Sigma 1 receptor: A novel therapeutic target in retinal disease.Prog Retin Eye Res. 2018 Nov;67:130-149. doi: 10.1016/j.preteyeres.2018.07.003. Epub 2018 Aug 1. Prog Retin Eye Res. 2018. PMID: 30075336 Free PMC article. Review.
-
The Role of Sigma1R in Mammalian Retina.Adv Exp Med Biol. 2017;964:267-284. doi: 10.1007/978-3-319-50174-1_18. Adv Exp Med Biol. 2017. PMID: 28315277 Review.
Cited by
-
Machine Learning Assistants Construct Oxidative Stress-Related Gene Signature and Discover Potential Therapy Targets for Acute Myeloid Leukemia.Oxid Med Cell Longev. 2022 Aug 22;2022:1507690. doi: 10.1155/2022/1507690. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 36046688 Free PMC article.
-
Suppression of cGMP-Dependent Photoreceptor Cytotoxicity With Mycophenolate Is Neuroprotective in Murine Models of Retinitis Pigmentosa.Invest Ophthalmol Vis Sci. 2020 Aug 3;61(10):25. doi: 10.1167/iovs.61.10.25. Invest Ophthalmol Vis Sci. 2020. PMID: 32785677 Free PMC article.
-
Hyperhomocysteinemia-induced death of retinal ganglion cells: The role of Müller glial cells and NRF2.Redox Biol. 2019 Jun;24:101199. doi: 10.1016/j.redox.2019.101199. Epub 2019 Apr 11. Redox Biol. 2019. PMID: 31026769 Free PMC article.
-
Neuroprotection of retinal ganglion cells by the sigma-1 receptor agonist pridopidine in models of experimental glaucoma.Sci Rep. 2021 Nov 9;11(1):21975. doi: 10.1038/s41598-021-01077-w. Sci Rep. 2021. PMID: 34753986 Free PMC article.
-
Sigma-1 Receptor in Retina: Neuroprotective Effects and Potential Mechanisms.Int J Mol Sci. 2022 Jul 8;23(14):7572. doi: 10.3390/ijms23147572. Int J Mol Sci. 2022. PMID: 35886921 Free PMC article. Review.
References
-
- Cuenca N, et al. Cellular responses following retinal injuries and therapeutic approaches for neurodegenerative diseases. Prog Retin Eye Res. 2014;43:17–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

