Chaperoning SNARE assembly and disassembly
- PMID: 27301672
- PMCID: PMC5471617
- DOI: 10.1038/nrm.2016.65
Chaperoning SNARE assembly and disassembly
Abstract
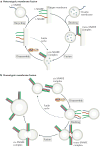
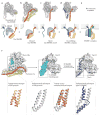
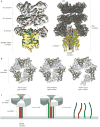
Intracellular membrane fusion is mediated in most cases by membrane-bridging complexes of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs). However, the assembly of such complexes in vitro is inefficient, and their uncatalysed disassembly is undetectably slow. Here, we focus on the cellular machinery that orchestrates assembly and disassembly of SNARE complexes, thereby regulating processes ranging from vesicle trafficking to organelle fusion to neurotransmitter release. Rapid progress is being made on many fronts, including the development of more realistic cell-free reconstitutions, the application of single-molecule biophysics, and the elucidation of X-ray and high-resolution electron microscopy structures of the SNARE assembly and disassembly machineries 'in action'.
Figures

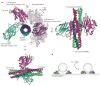
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

