Modeling altered T-cell development with induced pluripotent stem cells from patients with RAG1-dependent immune deficiencies
- PMID: 27301863
- PMCID: PMC4982452
- DOI: 10.1182/blood-2015-10-676304
Modeling altered T-cell development with induced pluripotent stem cells from patients with RAG1-dependent immune deficiencies
Abstract
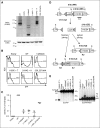
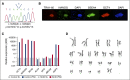
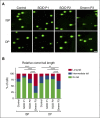
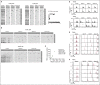
Primary immunodeficiency diseases comprise a group of heterogeneous genetic defects that affect immune system development and/or function. Here we use in vitro differentiation of human induced pluripotent stem cells (iPSCs) generated from patients with different recombination-activating gene 1 (RAG1) mutations to assess T-cell development and T-cell receptor (TCR) V(D)J recombination. RAG1-mutants from severe combined immunodeficient (SCID) patient cells showed a failure to sustain progression beyond the CD3(--)CD4(-)CD8(-)CD7(+)CD5(+)CD38(-)CD31(-/lo)CD45RA(+) stage of T-cell development to reach the CD3(-/+)CD4(+)CD8(+)CD7(+)CD5(+)CD38(+)CD31(+)CD45RA(-) stage. Despite residual mutant RAG1 recombination activity from an Omenn syndrome (OS) patient, similar impaired T-cell differentiation was observed, due to increased single-strand DNA breaks that likely occur due to heterodimers consisting of both an N-terminal truncated and a catalytically dead RAG1. Furthermore, deep-sequencing analysis of TCR-β (TRB) and TCR-α (TRA) rearrangements of CD3(-)CD4(+)CD8(-) immature single-positive and CD3(+)CD4(+)CD8(+) double-positive cells showed severe restriction of repertoire diversity with preferential usage of few Variable, Diversity, and Joining genes, and skewed length distribution of the TRB and TRA complementary determining region 3 sequences from SCID and OS iPSC-derived cells, whereas control iPSCs yielded T-cell progenitors with a broadly diversified repertoire. Finally, no TRA/δ excision circles (TRECs), a marker of TRA/δ locus rearrangements, were detected in SCID and OS-derived T-lineage cells, consistent with a pre-TCR block in T-cell development. This study compares human T-cell development of SCID vs OS patients, and elucidates important differences that help to explain the wide range of immunologic phenotypes that result from different mutations within the same gene of various patients.
© 2016 by The American Society of Hematology.
Figures


Comment in
-
Modeling altered human T-cell development.Blood. 2016 Aug 11;128(6):743-5. doi: 10.1182/blood-2016-06-723908. Blood. 2016. PMID: 27516425 No abstract available.
References
-
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(suppl):S45–S55. - PubMed
-
- Roth DB, Menetski JP, Nakajima PB, Bosma MJ, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70(6):983–991. - PubMed
-
- Fugmann SD. RAG1 and RAG2 in V(D)J recombination and transposition. Immunol Res. 2001;23(1):23–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

