Real-time 3D visualization of cellular rearrangements during cardiac valve formation
- PMID: 27302398
- PMCID: PMC4920173
- DOI: 10.1242/dev.133272
Real-time 3D visualization of cellular rearrangements during cardiac valve formation
Abstract
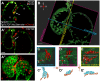
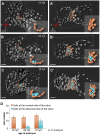
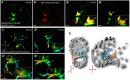
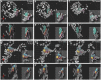
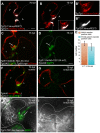
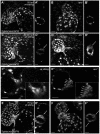
During cardiac valve development, the single-layered endocardial sheet at the atrioventricular canal (AVC) is remodeled into multilayered immature valve leaflets. Most of our knowledge about this process comes from examining fixed samples that do not allow a real-time appreciation of the intricacies of valve formation. Here, we exploit non-invasive in vivo imaging techniques to identify the dynamic cell behaviors that lead to the formation of the immature valve leaflets. We find that in zebrafish, the valve leaflets consist of two sets of endocardial cells at the luminal and abluminal side, which we refer to as luminal cells (LCs) and abluminal cells (ALCs), respectively. By analyzing cellular rearrangements during valve formation, we observed that the LCs and ALCs originate from the atrium and ventricle, respectively. Furthermore, we utilized Wnt/β-catenin and Notch signaling reporter lines to distinguish between the LCs and ALCs, and also found that cardiac contractility and/or blood flow is necessary for the endocardial expression of these signaling reporters. Thus, our 3D analyses of cardiac valve formation in zebrafish provide fundamental insights into the cellular rearrangements underlying this process.
Keywords: Apc; Cardiac valves; Endocardium; Live imaging; Notch; Wnt; Zebrafish.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures
References
-
- Asakawa K., Suster M. L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M. and Kawakami K. (2008). Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA 105, 1255-1260. 10.1073/pnas.0704963105 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

