Conserved 33-kb haplotype in the MHC class III region regulates chronic arthritis
- PMID: 27303036
- PMCID: PMC4932949
- DOI: 10.1073/pnas.1600567113
Conserved 33-kb haplotype in the MHC class III region regulates chronic arthritis
Abstract
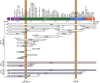
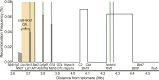
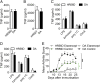
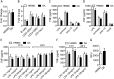
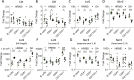
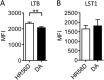
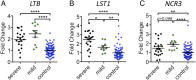
Genome-wide association studies have revealed many genetic loci associated with complex autoimmune diseases. In rheumatoid arthritis (RA), the MHC gene HLA-DRB1 is the strongest candidate predicting disease development. It has been suggested that other immune-regulating genes in the MHC contribute to the disease risk, but this contribution has been difficult to show because of the strong linkage disequilibrium within the MHC. We isolated genomic regions in the form of congenic fragments in rats to test whether there are additional susceptibility loci in the MHC. By both congenic mapping in inbred strains and SNP typing in wild rats, we identified a conserved, 33-kb large haplotype Ltab-Ncr3 in the MHC-III region, which regulates the onset, severity, and chronicity of arthritis. The Ltab-Ncr3 haplotype consists of five polymorphic immunoregulatory genes: Lta (lymphotoxin-α), Tnf, Ltb (lymphotoxin-β), Lst1 (leukocyte-specific transcript 1), and Ncr3 (natural cytotoxicity-triggering receptor 3). Significant correlation in the expression of the Ltab-Ncr3 genes suggests that interaction of these genes may be important in keeping these genes clustered together as a conserved haplotype. We studied the arthritis association and the spliceo-transcriptome of four different Ltab-Ncr3 haplotypes and showed that higher Ltb and Ncr3 expression, lower Lst1 expression, and the expression of a shorter splice variant of Lst1 correlate with reduced arthritis severity in rats. Interestingly, patients with mild RA also showed higher NCR3 expression and lower LST1 expression than patients with severe RA. These data demonstrate the importance of a conserved haplotype in the regulation of complex diseases such as arthritis.
Keywords: arthritis; congenic mapping; haplotype; inflammation; major histocompatibility complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Comment in
-
Cis-eQTLs regulate reduced LST1 gene and NCR3 gene expression and contribute to increased autoimmune disease risk.Proc Natl Acad Sci U S A. 2016 Oct 18;113(42):E6321-E6322. doi: 10.1073/pnas.1614369113. Epub 2016 Oct 11. Proc Natl Acad Sci U S A. 2016. PMID: 27729515 Free PMC article. No abstract available.
References
-
- Messemaker TC, Huizinga TW, Kurreeman F. Immunogenetics of rheumatoid arthritis: Understanding functional implications. J Autoimmun. 2015;64:74–81. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

