RNA-Seq Analysis of Differential Gene Expression Responding to Different Rhizobium Strains in Soybean (Glycine max) Roots
- PMID: 27303417
- PMCID: PMC4885319
- DOI: 10.3389/fpls.2016.00721
RNA-Seq Analysis of Differential Gene Expression Responding to Different Rhizobium Strains in Soybean (Glycine max) Roots
Abstract
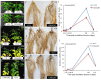
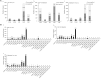
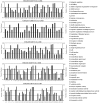
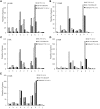
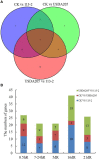
The root nodule symbiosis (RNS) between legume plants and rhizobia is the most efficient and productive source of nitrogen fixation, and has critical importance in agriculture and mesology. Soybean (Glycine max), one of the most important legume crops in the world, establishes a nitrogen-fixing symbiosis with different types of rhizobia, and the efficiency of symbiotic nitrogen fixation in soybean greatly depends on the symbiotic host-specificity. Although, it has been reported that rhizobia use surface polysaccharides, secretion proteins of the type-three secretion systems and nod factors to modulate host range, the host control of nodulation specificity remains poorly understood. In this report, the soybean roots of two symbiotic systems (Bradyrhizobium japonicum strain 113-2-soybean and Sinorhizobium fredii USDA205-soybean)with notable different nodulation phenotypes and the control were studied at five different post-inoculation time points (0.5, 7-24 h, 5, 16, and 21 day) by RNA-seq (Quantification). The results of qPCR analysis of 11 randomly-selected genes agreed with transcriptional profile data for 136 out of 165 (82.42%) data points and quality assessment showed that the sequencing library is of quality and reliable. Three comparisons (control vs. 113-2, control vs. USDA205 and USDA205 vs. 113-2) were made and the differentially expressed genes (DEGs) between them were analyzed. The number of DEGs at 16 days post-inoculation (dpi) was the highest in the three comparisons, and most of the DEGs in USDA205 vs. 113-2 were found at 16 dpi and 21 dpi. 44 go function terms in USDA205 vs. 113-2 were analyzed to evaluate the potential functions of the DEGs, and 10 important KEGG pathway enrichment terms were analyzed in the three comparisons. Some important genes induced in response to different strains (113-2 and USDA205) were identified and analyzed, and these genes primarily encoded soybean resistance proteins, NF-related proteins, nodulins and immunity defense proteins, as well as proteins involving flavonoids/flavone/flavonol biosynthesis and plant-pathogen interaction. Besides, 189 candidate genes are largely expressed in roots and\or nodules. The DEGs uncovered in this study provides molecular candidates for better understanding the mechanisms of symbiotic host-specificity and explaining the different symbiotic effects between soybean roots inoculated with different strains (113-2 and USDA205).
Keywords: RNA-seq; Soybean; different nodulation phenotypes; differential gene expression responding; symbiotic specificity.
Figures
Similar articles
-
RNA-Seq analysis of mung bean (Vigna radiata L.) roots shows differential gene expression and predicts regulatory pathways responding to taxonomically different rhizobia.Microbiol Res. 2023 Oct;275:127451. doi: 10.1016/j.micres.2023.127451. Epub 2023 Jul 12. Microbiol Res. 2023. PMID: 37478540
-
RNA-Seq analysis of nodule development at five different developmental stages of soybean (Glycine max) inoculated with Bradyrhizobium japonicum strain 113-2.Sci Rep. 2017 Feb 7;7:42248. doi: 10.1038/srep42248. Sci Rep. 2017. PMID: 28169364 Free PMC article.
-
Classical Soybean (Glycine max (L.) Merr) Symbionts, Sinorhizobium fredii USDA191 and Bradyrhizobium diazoefficiens USDA110, Reveal Contrasting Symbiotic Phenotype on Pigeon Pea (Cajanus cajan (L.) Millsp).Int J Mol Sci. 2019 Mar 3;20(5):1091. doi: 10.3390/ijms20051091. Int J Mol Sci. 2019. PMID: 30832430 Free PMC article.
-
Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean.J Biotechnol. 2011 Aug 20;155(1):11-9. doi: 10.1016/j.jbiotec.2011.03.016. Epub 2011 Mar 30. J Biotechnol. 2011. PMID: 21458507 Review.
-
Nodulation outer proteins: double-edged swords of symbiotic rhizobia.Biochem J. 2015 Sep 15;470(3):263-74. doi: 10.1042/BJ20150518. Biochem J. 2015. PMID: 26341483 Review.
Cited by
-
Whole-Genome Sequencing of Bradyrhizobium diazoefficiens 113-2 and Comparative Genomic Analysis Provide Molecular Insights Into Species Specificity and Host Specificity.Front Microbiol. 2020 Nov 16;11:576800. doi: 10.3389/fmicb.2020.576800. eCollection 2020. Front Microbiol. 2020. PMID: 33329441 Free PMC article.
-
Aberrant Meiotic Prophase I Leads to Genic Male Sterility in the Novel TE5A Mutant of Brassica napus.Sci Rep. 2016 Sep 27;6:33955. doi: 10.1038/srep33955. Sci Rep. 2016. PMID: 27670217 Free PMC article.
-
Genome-wide survey of soybean papain-like cysteine proteases and their expression analysis in root nodule symbiosis.BMC Plant Biol. 2020 Nov 12;20(1):517. doi: 10.1186/s12870-020-02725-5. BMC Plant Biol. 2020. PMID: 33183238 Free PMC article.
-
Genome-Wide Identification and Classification of Soybean C2H2 Zinc Finger Proteins and Their Expression Analysis in Legume-Rhizobium Symbiosis.Front Microbiol. 2018 Feb 6;9:126. doi: 10.3389/fmicb.2018.00126. eCollection 2018. Front Microbiol. 2018. PMID: 29467740 Free PMC article.
-
Transcriptome analysis of soybean (Glycine max) root genes differentially expressed in rhizobial, arbuscular mycorrhizal, and dual symbiosis.J Plant Res. 2019 Jul;132(4):541-568. doi: 10.1007/s10265-019-01117-7. Epub 2019 Jun 5. J Plant Res. 2019. PMID: 31165947
References
-
- Annapurna K., Krishnan H. B. (2003). Molecular aspects of soybean cultivar-specific nodulation by Sinorhizobium fredii USDA257. Indian J. Exp. Biol. 41, 1114–1123. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

