Glycosylation Is a Major Regulator of Phenylpropanoid Availability and Biological Activity in Plants
- PMID: 27303427
- PMCID: PMC4880792
- DOI: 10.3389/fpls.2016.00735
Glycosylation Is a Major Regulator of Phenylpropanoid Availability and Biological Activity in Plants
Abstract
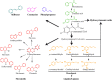
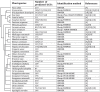
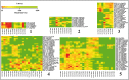
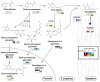
The phenylpropanoid pathway in plants is responsible for the biosynthesis of a huge amount of secondary metabolites derived from phenylalanine and tyrosine. Both flavonoids and lignins are synthesized at the end of this very diverse metabolic pathway, as well as many intermediate molecules whose precise biological functions remain largely unknown. The diversity of these molecules can be further increased under the action of UDP-glycosyltransferases (UGTs) leading to the production of glycosylated hydroxycinnamates and related aldehydes, alcohols and esters. Glycosylation can change phenylpropanoid solubility, stability and toxic potential, as well as influencing compartmentalization and biological activity. (De)-glycosylation therefore represents an extremely important regulation point in phenylpropanoid homeostasis. In this article we review recent knowledge on the enzymes involved in regulating phenylpropanoid glycosylation status and availability in different subcellular compartments. We also examine the potential link between monolignol glycosylation and lignification by exploring co-expression of lignin biosynthesis genes and phenolic (de)glycosylation genes. Of the different biological roles linked with their particular chemical properties, phenylpropanoids are often correlated with the plant's stress management strategies that are also regulated by glycosylation. UGTs can for instance influence the resistance of plants during infection by microorganisms and be involved in the mechanisms related to environmental changes. The impact of flavonoid glycosylation on the color of flowers, leaves, seeds and fruits will also be discussed. Altogether this paper underlies the fact that glycosylation and deglycosylation are powerful mechanisms allowing plants to regulate phenylpropanoid localisation, availability and biological activity.
Keywords: UDP-glycosyltransferase; beta-glucosidase; compartmentalization; flavonoids; glycosylation; lignin; phenylpropanoids.
Figures
References
-
- Ahrazem O., Rubio-Moraga A., Trapero-Mozos A., Climent M. F., Gomez-Cadenas A., Gomez-Gomez L. (2015). Ectopic expression of a stress-inducible glycosyltransferase from saffron enhances salt and oxidative stress tolerance in Arabidopsis while alters anchor root formation. Plant Sci 234, 60–73. 10.1016/j.plantsci.2015.02.004 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

