Novel Immunomodulatory Flagellin-Like Protein FlaC in Campylobacter jejuni and Other Campylobacterales
- PMID: 27303676
- PMCID: PMC4863622
- DOI: 10.1128/mSphere.00028-15
Novel Immunomodulatory Flagellin-Like Protein FlaC in Campylobacter jejuni and Other Campylobacterales
Abstract
The human diarrheal pathogens Campylobacter jejuni and Campylobacter coli interfere with host innate immune signaling by different means, and their flagellins, FlaA and FlaB, have a low intrinsic property to activate the innate immune receptor Toll-like receptor 5 (TLR5). We have investigated here the hypothesis that the unusual secreted, flagellin-like molecule FlaC present in C. jejuni, C. coli, and other Campylobacterales might activate cells via TLR5 and interact with TLR5. FlaC shows striking sequence identity in its D1 domains to TLR5-activating flagellins of other bacteria, such as Salmonella, but not to nonstimulating Campylobacter flagellins. We overexpressed and purified FlaC and tested its immunostimulatory properties on cells of human and chicken origin. Treatment of cells with highly purified FlaC resulted in p38 activation. FlaC directly interacted with TLR5. Preincubation with FlaC decreased the responsiveness of chicken and human macrophage-like cells toward the bacterial TLR4 agonist lipopolysaccharide (LPS), suggesting that FlaC mediates cross-tolerance. C. jejuni flaC mutants induced an increase of cell responses in comparison to those of the wild type, which was suppressed by genetic complementation. Supplementing excess purified FlaC likewise reduced the cellular response to C. jejuni. In vivo, the administration of ultrapure FlaC led to a decrease in cecal interleukin 1β (IL-1β) expression and a significant change of the cecal microbiota in chickens. We propose that Campylobacter spp. have evolved a novel type of secreted immunostimulatory flagellin-like effector in order to specifically modulate host responses, for example toward other pattern recognition receptor (PRR) ligands, such as LPS. IMPORTANCE Flagellins not only are important for bacterial motility but are major bacterial proteins that can modulate host responses via Toll-like receptor 5 (TLR5) or other pattern recognition receptors. Campylobacterales colonizing the intestinal tracts of different host species harbor a gene coding for an unusual flagellin, FlaC, that is not involved in motility but is secreted and possesses a chimeric amino acid sequence composed of TLR5-activating and non-TLR5-activating flagellin sequences. Campylobacter jejuni FlaC activates cells to increase in cytokine expression in chicken and human cells, promotes cross-tolerance to TLR4 ligands, and alters chicken cecal microbiota. We propose that FlaC is a secreted effector flagellin that has specifically evolved to modulate the immune response in the intestinal tract in the presence of the resident microbiota and may contribute to bacterial persistence. The results also strengthen the role of the flagellar type III apparatus as a functional secretion system for bacterial effector proteins.
Keywords: Campylobacter; TLR5; flagellin; host-pathogen interaction; immune response.
Figures

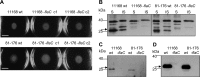
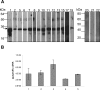
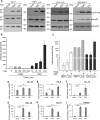



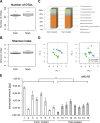
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
