Manipulation of the Gut Microbiota Reveals Role in Colon Tumorigenesis
- PMID: 27303681
- PMCID: PMC4863627
- DOI: 10.1128/mSphere.00001-15
Manipulation of the Gut Microbiota Reveals Role in Colon Tumorigenesis
Abstract
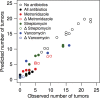
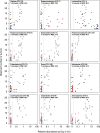
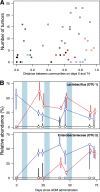
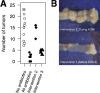
There is growing evidence that individuals with colonic adenomas and carcinomas harbor a distinct microbiota. Alterations to the gut microbiota may allow the outgrowth of bacterial populations that induce genomic mutations or exacerbate tumor-promoting inflammation. In addition, it is likely that the loss of key bacterial populations may result in the loss of protective functions that are normally provided by the microbiota. We explored the role of the gut microbiota in colon tumorigenesis by using an inflammation-based murine model. We observed that perturbing the microbiota with different combinations of antibiotics reduced the number of tumors at the end of the model. Using the random forest machine learning algorithm, we successfully modeled the number of tumors that developed over the course of the model on the basis of the initial composition of the microbiota. The timing of antibiotic treatment was an important determinant of tumor outcome, as colon tumorigenesis was arrested by the use of antibiotics during the early inflammation period of the murine model. Together, these results indicate that it is possible to predict colon tumorigenesis on the basis of the composition of the microbiota and that altering the gut microbiota can alter the course of tumorigenesis. IMPORTANCE Mounting evidence indicates that alterations to the gut microbiota, the complex community of bacteria that inhabits the gastrointestinal tract, are strongly associated with the development of colorectal cancer. We used antibiotic perturbations to a murine model of inflammation-driven colon cancer to generate eight starting communities that resulted in various severities of tumorigenesis. Furthermore, we were able to quantitatively predict the final number of tumors on the basis of the initial composition of the gut microbiota. These results further bolster the evidence that the gut microbiota is involved in mediating the development of colorectal cancer. As a final proof of principle, we showed that perturbing the gut microbiota in the midst of tumorigenesis could halt the formation of additional tumors. Together, alteration of the gut microbiota may be a useful therapeutic approach to preventing and altering the trajectory of colorectal cancer.
Keywords: 16S rRNA gene sequencing; azoxymethane; colorectal cancer; dextran sodium sulfate; microbial ecology; microbiome; murine models.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
