Characterization of Five Novel Brevibacillus Bacteriophages and Genomic Comparison of Brevibacillus Phages
- PMID: 27304881
- PMCID: PMC4909266
- DOI: 10.1371/journal.pone.0156838
Characterization of Five Novel Brevibacillus Bacteriophages and Genomic Comparison of Brevibacillus Phages
Abstract
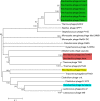
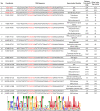
Brevibacillus laterosporus is a spore-forming bacterium that causes a secondary infection in beehives following European Foulbrood disease. To better understand the contributions of Brevibacillus bacteriophages to the evolution of their hosts, five novel phages (Jenst, Osiris, Powder, SecTim467, and Sundance) were isolated and characterized. When compared with the five Brevibacillus phages currently in NCBI, these phages were assigned to clusters based on whole genome and proteome synteny. Powder and Osiris, both myoviruses, were assigned to the previously described Jimmer-like cluster. SecTim467 and Jenst, both siphoviruses, formed a novel phage cluster. Sundance, a siphovirus, was assigned as a singleton phage along with the previously isolated singleton, Emery. In addition to characterizing the basic relationships between these phages, several genomic features were observed. A motif repeated throughout phages Jenst and SecTim467 was frequently upstream of genes predicted to function in DNA replication, nucleotide metabolism, and transcription, suggesting transcriptional co-regulation. In addition, paralogous gene pairs that encode a putative transcriptional regulator were identified in four Brevibacillus phages. These paralogs likely evolved to bind different DNA sequences due to variation at amino acid residues predicted to bind specific nucleotides. Finally, a putative transposable element was identified in SecTim467 and Sundance that carries genes homologous to those found in Brevibacillus chromosomes. Remnants of this transposable element were also identified in phage Jenst. These discoveries provide a greater understanding of the diversity of phages, their behavior, and their evolutionary relationships to one another and to their host. In addition, they provide a foundation with which further Brevibacillus phages can be compared.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

