Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi
- PMID: 27304955
- PMCID: PMC4926463
- DOI: 10.3390/ijms17060930
Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi
Abstract
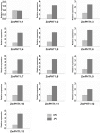
The Phosphate Transporter1 (PHT1) family of genes plays pivotal roles in the uptake of inorganic phosphate from soils. However, there is no comprehensive report on the PHT1 family in Zea mays based on the whole genome. In the present study, a total of 13 putative PHT1 genes (ZmPHT1;1 to 13) were identified in the inbred line B73 genome by bioinformatics methods. Then, their function was investigated by a yeast PHO84 mutant complementary experiment and qRT-PCR. Thirteen ZmPHT1 genes distributed on six chromosomes (1, 2, 5, 7, 8 and 10) were divided into two paralogues (Class A and Class B). ZmPHT1;1/ZmPHT1;9 and ZmPHT1;9/ZmPHT1;13 are produced from recent segmental duplication events. ZmPHT1;1/ZmPHT1;13 and ZmPHT1;8/ZmPHT1;10 are produced from early segmental duplication events. All 13 putative ZmPHT1s can completely or partly complement the yeast Pi-uptake mutant, and they were obviously induced in maize under low Pi conditions, except for ZmPHT1;1 (p < 0.01), indicating that the overwhelming majority of ZmPHT1 genes can respond to a low Pi condition. ZmPHT1;2, ZmPHT1;4, ZmPHT1;6, ZmPHT1;7, ZmPHT1;9 and ZmPHT1;11 were up-regulated by arbuscular mycorrhizal fungi (AMF), implying that these genes might participate in mediating Pi absorption and/or transport. Analysis of the promoters revealed that the MYCS and P1BS element are widely distributed on the region of different AMF-inducible ZmPHT1 promoters. In light of the above results, five of 13 ZmPHT1 genes were newly-identified AMF-inducible high-affinity phosphate transporters in the maize genome. Our results will lay a foundation for better understanding the PHT1 family evolution and the molecular mechanisms of inorganic phosphate transport under AMF inoculation.
Keywords: AMF-inducible; Zea mays; evolution; expression; phosphate transporters; promoter.
Figures








References
-
- Van Der Heijden M.G.A., Streitwolf-Engel R., Riedl R., Siegrist S., Neudecker A., Ineichen K., Boller T., Wiemken A., Sanders I.R. The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland. New Phytol. 2006;172:739–752. doi: 10.1111/j.1469-8137.2006.01862.x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

