Qualitative dynamics semantics for SBGN process description
- PMID: 27306057
- PMCID: PMC4910245
- DOI: 10.1186/s12918-016-0285-0
Qualitative dynamics semantics for SBGN process description
Abstract
Background: Qualitative dynamics semantics provide a coarse-grain modeling of networks dynamics by abstracting away kinetic parameters. They allow to capture general features of systems dynamics, such as attractors or reachability properties, for which scalable analyses exist. The Systems Biology Graphical Notation Process Description language (SBGN-PD) has become a standard to represent reaction networks. However, no qualitative dynamics semantics taking into account all the main features available in SBGN-PD had been proposed so far.
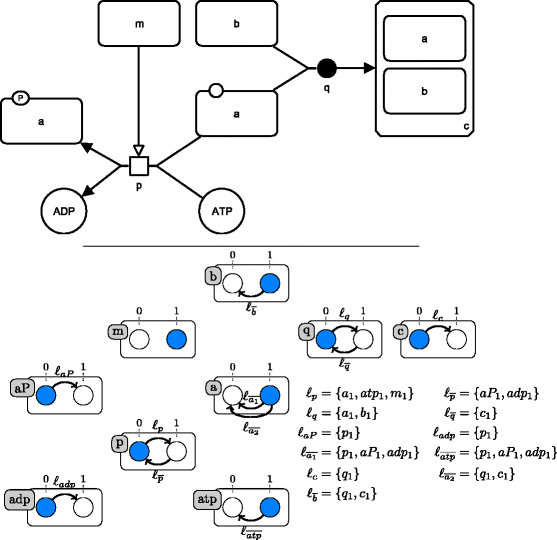
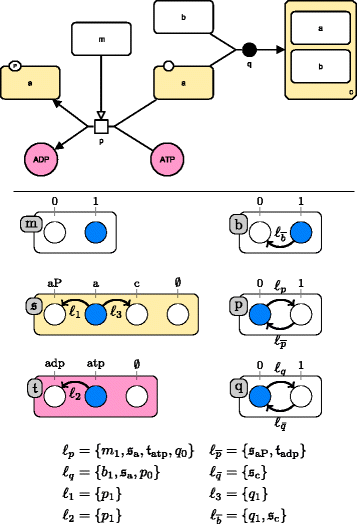
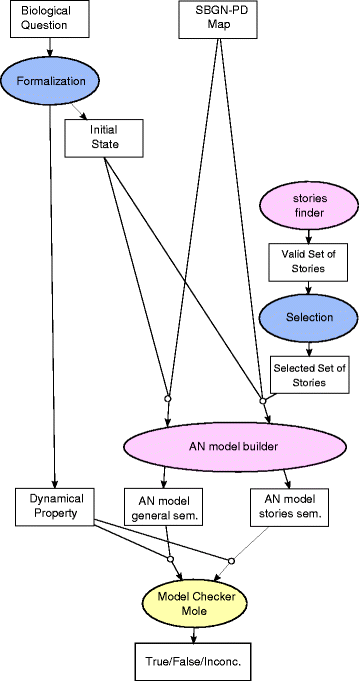
Results: We propose two qualitative dynamics semantics for SBGN-PD reaction networks, namely the general semantics and the stories semantics, that we formalize using asynchronous automata networks. While the general semantics extends standard Boolean semantics of reaction networks by taking into account all the main features of SBGN-PD, the stories semantics allows to model several molecules of a network by a unique variable. The obtained qualitative models can be checked against dynamical properties and therefore validated with respect to biological knowledge. We apply our framework to reason on the qualitative dynamics of a large network (more than 200 nodes) modeling the regulation of the cell cycle by RB/E2F.
Conclusion: The proposed semantics provide a direct formalization of SBGN-PD networks in dynamical qualitative models that can be further analyzed using standard tools for discrete models. The dynamics in stories semantics have a lower dimension than the general one and prune multiple behaviors (which can be considered as spurious) by enforcing the mutual exclusiveness between the activity of different nodes of a same story. Overall, the qualitative semantics for SBGN-PD allow to capture efficiently important dynamical features of reaction network models and can be exploited to further refine them.
Keywords: Automata networks; Modeling of dynamics; Qualitative dynamics; Reaction networks; SBGN-PD.
Figures







Similar articles
-
Systems Biology Graphical Notation: Process Description language Level 1 Version 1.3.J Integr Bioinform. 2015 Sep 4;12(2):263. doi: 10.2390/biecoll-jib-2015-263. J Integr Bioinform. 2015. PMID: 26528561
-
Systems Biology Graphical Notation: Activity Flow language Level 1 Version 1.2.J Integr Bioinform. 2015 Sep 4;12(2):265. doi: 10.2390/biecoll-jib-2015-265. J Integr Bioinform. 2015. PMID: 26528563
-
Systems Biology Graphical Notation: Entity Relationship language Level 1 Version 2.J Integr Bioinform. 2015 Sep 4;12(2):264. doi: 10.2390/biecoll-jib-2015-264. J Integr Bioinform. 2015. PMID: 26528562
-
Systems Biology Graphical Notation: Process Description language Level 1 Version 2.0.J Integr Bioinform. 2019 Jun 13;16(2):20190022. doi: 10.1515/jib-2019-0022. J Integr Bioinform. 2019. PMID: 31199769 Free PMC article. Review.
-
Biochemical modeling with Systems Biology Graphical Notation.Drug Discov Today. 2010 May;15(9-10):365-70. doi: 10.1016/j.drudis.2010.02.012. Epub 2010 Mar 6. Drug Discov Today. 2010. PMID: 20211756 Review.
Cited by
-
A detailed map of coupled circadian clock and cell cycle with qualitative dynamics validation.BMC Bioinformatics. 2021 May 11;22(1):240. doi: 10.1186/s12859-021-04158-9. BMC Bioinformatics. 2021. PMID: 33975535 Free PMC article.
-
Discrete modeling for integration and analysis of large-scale signaling networks.PLoS Comput Biol. 2022 Jun 13;18(6):e1010175. doi: 10.1371/journal.pcbi.1010175. eCollection 2022 Jun. PLoS Comput Biol. 2022. PMID: 35696426 Free PMC article.
-
A logic-based method to build signaling networks and propose experimental plans.Sci Rep. 2018 May 18;8(1):7830. doi: 10.1038/s41598-018-26006-2. Sci Rep. 2018. PMID: 29777117 Free PMC article.
References
-
- KEGG Pathway Database. http://www.genome.jp/kegg/pathway.html%23metabolism. Accessed 2016-02-08.
-
- ACSN - Atlas of Cancer Signalling Networks. https://acsn.curie.fr. Accessed 2016-02-08.
-
- Calzone L, Gelay A, Zinovyev A, Radvanyi F, Barillot E. A comprehensive modular map of molecular interactions in RB/E2F pathway. Mol Syst Biol. 2008; 4(1). http://msb.embopress.org/content/4/1/0174.export. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

