Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A
- PMID: 27306458
- PMCID: PMC4912631
- DOI: 10.1038/ncomms11952
Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A
Abstract
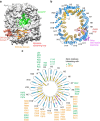
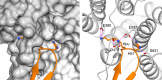
Xpo4 is a bidirectional nuclear transport receptor that mediates nuclear export of eIF5A and Smad3 as well as import of Sox2 and SRY. How Xpo4 recognizes such a variety of cargoes is as yet unknown. Here we present the crystal structure of the RanGTP·Xpo4·eIF5A export complex at 3.2 Å resolution. Xpo4 has a similar structure as CRM1, but the NES-binding site is occluded, and a new interaction site evolved that recognizes both globular domains of eIF5A. eIF5A contains hypusine, a unique amino acid with two positive charges, which is essential for cell viability and eIF5A function in translation. The hypusine docks into a deep, acidic pocket of Xpo4 and is thus a critical element of eIF5A's complex export signature. This further suggests that Xpo4 recognizes other cargoes differently, and illustrates how Xpo4 suppresses - in a chaperone-like manner - undesired interactions of eIF5A inside nuclei.
Figures





References
-
- Sloan K. E., Gleizes P. E. & Bohnsack M. T. Nucleocytoplasmic transport of RNAs and RNA-protein complexes. J. Mol. Biol. 428, 2040–2059 (2015). - PubMed
-
- Hurt E. & Beck M. Towards understanding nuclear pore complex architecture and dynamics in the age of integrative structural analysis. Curr. Opin. Cell Biol. 34, 31–38 (2015). - PubMed
-
- Schmidt H. B. & Görlich D. Transport Selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 41, 46–61 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

