The Rickettsia type IV secretion system: unrealized complexity mired by gene family expansion
- PMID: 27307105
- PMCID: PMC5505475
- DOI: 10.1093/femspd/ftw058
The Rickettsia type IV secretion system: unrealized complexity mired by gene family expansion
Abstract
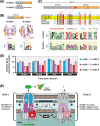
Many prokaryotes utilize type IV secretion systems (T4SSs) to translocate substrates (e.g. nucleoprotein, DNA, protein) across the cell envelope, and/or to elaborate surface structures (i.e. pili or adhesins). Among eight distinct T4SS classes, P-T4SSs are typified by the Agrobacterium tumefaciens vir T4SS, which is comprised of 12 scaffold components (VirB1-VirB11, VirD4). While most P-T4SSs include all 12 Vir proteins, some differ from the vir archetype by either containing additional scaffold components not analogous to Vir proteins or lacking one or more of the Vir proteins. In a special case, the Rickettsiales vir homolog (rvh) P-T4SS comprises unprecedented gene family expansion. rvh contains three families of gene duplications (rvhB9, rvhB8, rvhB4): RvhB9,8,4-I are conserved relative to equivalents in other P-T4SSs, while RvhB9,8,4-II have evolved atypical features that deviate substantially from other homologs. Furthermore, rvh contains five VirB6-like genes (rvhB6a-e), which are tandemly arrayed and contain large N- and C-terminal extensions. Our work herein focuses on the complexity underpinned by rvh gene family expansion. Furthermore, we describe an RvhB10 insertion, which occurs in a region that forms the T4SS pore. The significance of these curious properties to rvh structure and function is evaluated, shedding light on a highly complex T4SS.
Keywords: Rickettsia; Rickettsiales vir homolog; gene duplication; obligate intracellular bacteria; pathogenesis; type IV secretion system.
© FEMS 2016. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



Similar articles
-
Structural Insight into How Bacteria Prevent Interference between Multiple Divergent Type IV Secretion Systems.mBio. 2015 Dec 8;6(6):e01867-15. doi: 10.1128/mBio.01867-15. mBio. 2015. PMID: 26646013 Free PMC article.
-
An anomalous type IV secretion system in Rickettsia is evolutionarily conserved.PLoS One. 2009;4(3):e4833. doi: 10.1371/journal.pone.0004833. Epub 2009 Mar 12. PLoS One. 2009. PMID: 19279686 Free PMC article.
-
Membrane and core periplasmic Agrobacterium tumefaciens virulence Type IV secretion system components localize to multiple sites around the bacterial perimeter during lateral attachment to plant cells.mBio. 2011 Oct 25;2(6):e00218-11. doi: 10.1128/mBio.00218-11. Print 2011. mBio. 2011. PMID: 22027007 Free PMC article.
-
Phylogenomics reveals a diverse Rickettsiales type IV secretion system.Infect Immun. 2010 May;78(5):1809-23. doi: 10.1128/IAI.01384-09. Epub 2010 Feb 22. Infect Immun. 2010. PMID: 20176788 Free PMC article. Review.
-
Biological and Structural Diversity of Type IV Secretion Systems.Microbiol Spectr. 2019 Mar;7(2):10.1128/microbiolspec.psib-0012-2018. doi: 10.1128/microbiolspec.PSIB-0012-2018. Microbiol Spectr. 2019. PMID: 30953428 Free PMC article. Review.
Cited by
-
Involvement of Pore Formation and Osmotic Lysis in the Rapid Killing of Gamma Interferon-Pretreated C166 Endothelial Cells by Rickettsia prowazekii.Trop Med Infect Dis. 2022 Aug 1;7(8):163. doi: 10.3390/tropicalmed7080163. Trop Med Infect Dis. 2022. PMID: 36006255 Free PMC article.
-
Deianiraea, an extracellular bacterium associated with the ciliate Paramecium, suggests an alternative scenario for the evolution of Rickettsiales.ISME J. 2019 Sep;13(9):2280-2294. doi: 10.1038/s41396-019-0433-9. Epub 2019 May 9. ISME J. 2019. PMID: 31073215 Free PMC article.
-
Subversion of host cell signaling: The arsenal of Rickettsial species.Front Cell Infect Microbiol. 2022 Oct 31;12:995933. doi: 10.3389/fcimb.2022.995933. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36389139 Free PMC article. Review.
-
Cell-selective proteomics reveal novel effectors secreted by an obligate intracellular bacterial pathogen.Nat Commun. 2024 Jul 18;15(1):6073. doi: 10.1038/s41467-024-50493-9. Nat Commun. 2024. PMID: 39025857 Free PMC article.
-
Addictive manipulation: a perspective on the role of reproductive parasitism in the evolution of bacteria-eukaryote symbioses.Biol Lett. 2024 Sep;20(9):20240310. doi: 10.1098/rsbl.2024.0310. Epub 2024 Sep 18. Biol Lett. 2024. PMID: 39288812 Review.
References
-
- Andersson SG, Zomorodipour A, Andersson JO et al. . The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 1998;396:133–40 - PubMed
-
- Andrieux L, Bourg G, Pirone A et al. . A single amino acid change in the transmembrane domain of the VirB8 protein affects dimerization, interaction with VirB10 and Brucella suis virulence. FEBS Lett 2011;585:2431–6 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

