Posttranslational control of HuR function
- PMID: 27307117
- PMCID: PMC5607777
- DOI: 10.1002/wrna.1372
Posttranslational control of HuR function
Abstract
The RNA-binding protein HuR (human antigen R) associates with numerous transcripts, coding and noncoding, and controls their splicing, localization, stability, and translation. Through its regulation of target transcripts, HuR has been implicated in cellular events including proliferation, senescence, differentiation, apoptosis, and the stress and immune responses. In turn, HuR influences processes such as cancer and inflammation. HuR function is primarily regulated through posttranslational modifications that alter its subcellular localization and its ability to bind target RNAs; such modifications include phosphorylation, methylation, ubiquitination, NEDDylation, and proteolytic cleavage. In this review, we describe the modifications that impact upon HuR function on gene expression programs and disease states. WIREs RNA 2017, 8:e1372. doi: 10.1002/wrna.1372 For further resources related to this article, please visit the WIREs website.
Published 2016. This article is a U.S. Government work and is in the public domain in the USA.
Conflict of interest statement
Conflict of interest: The authors have declared no conflicts of interest for this article.
Figures
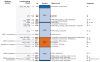
References
-
- Uy R, Wold F. Posttranslational covalent modification of proteins. Science. 1977;198:890–896. - PubMed
-
- Xin F, Radivojac P. Post-translational modifications induce significant yet not extreme changes to protein structure. Bioinformatics. 2012;28:2905–2913. - PubMed
-
- Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. - PubMed
-
- Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. - PubMed
-
- Martin L, Latypova X, Terro F. Post-translational modifications of τ protein: implications for Alzheimer’s disease. Neurochem Int. 2011;58:458–471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

