Dynamic DNA Methylation Regulates Levodopa-Induced Dyskinesia
- PMID: 27307239
- PMCID: PMC5015786
- DOI: 10.1523/JNEUROSCI.0683-16.2016
Dynamic DNA Methylation Regulates Levodopa-Induced Dyskinesia
Abstract
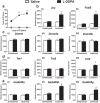
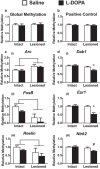
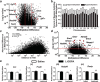
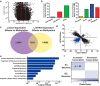
Levodopa-induced dyskinesia (LID) is a persistent behavioral sensitization that develops after repeated levodopa (l-DOPA) exposure in Parkinson disease patients. LID is a consequence of sustained changes in the transcriptional behavior of striatal neurons following dopaminergic stimulation. In neurons, transcriptional regulation through dynamic DNA methylation has been shown pivotal to many long-term behavioral modifications; however, its role in LID has not yet been explored. Using a rodent model, we show LID development leads to the aberrant expression of DNA demethylating enzymes and locus-specific changes to DNA methylation at the promoter regions of genes aberrantly transcribed following l-DOPA treatment. Looking for dynamic DNA methylation in LID genome-wide, we used reduced representation bisulfite sequencing and found an extensive reorganization of the dorsal striatal methylome. LID development led to significant demethylation at many important regulatory areas of aberrantly transcribed genes. We used pharmacologic treatments that alter DNA methylation bidirectionally and found them able to modulate dyskinetic behaviors. Together, these findings demonstrate that l-DOPA induces widespread changes to striatal DNA methylation and that these modifications are required for the development and maintenance of LID.
Significance statement: Levodopa-induced dyskinesia (LID) develops after repeated levodopa (l-DOPA) exposure in Parkinson disease patients and remains one of the primary obstacles to effective treatment. LID behaviors are a consequence of striatal neuron sensitization due to sustained changes in transcriptional behavior; however, the mechanisms responsible for the long-term maintenance of this cellular priming remain uncertain. Regulation of dynamic DNA methylation has been shown pivotal to the maintenance of several long-term behavioral modifications, yet its role in LID has not yet been explored. In this work, we report a pivotal role for the reorganization of DNA methylation in the development of LID and show that modification of DNA methylation may be a novel therapeutic target for use in preventing or reversing dyskinetic behaviors.
Keywords: DNA methylation; dyskinesia; l-DOPA.
Copyright © 2016 the authors 0270-6474/16/366514-11$15.00/0.
Figures

References
-
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, Vinson C, Stamatoyannopoulos JA, Hager GL. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
