Structures of trehalose-6-phosphate phosphatase from pathogenic fungi reveal the mechanisms of substrate recognition and catalysis
- PMID: 27307435
- PMCID: PMC4932955
- DOI: 10.1073/pnas.1601774113
Structures of trehalose-6-phosphate phosphatase from pathogenic fungi reveal the mechanisms of substrate recognition and catalysis
Abstract
Trehalose is a disaccharide essential for the survival and virulence of pathogenic fungi. The biosynthesis of trehalose requires trehalose-6-phosphate synthase, Tps1, and trehalose-6-phosphate phosphatase, Tps2. Here, we report the structures of the N-terminal domain of Tps2 (Tps2NTD) from Candida albicans, a transition-state complex of the Tps2 C-terminal trehalose-6-phosphate phosphatase domain (Tps2PD) bound to BeF3 and trehalose, and catalytically dead Tps2PD(D24N) from Cryptococcus neoformans bound to trehalose-6-phosphate (T6P). The Tps2NTD closely resembles the structure of Tps1 but lacks any catalytic activity. The Tps2PD-BeF3-trehalose and Tps2PD(D24N)-T6P complex structures reveal a "closed" conformation that is effected by extensive interactions between each trehalose hydroxyl group and residues of the cap and core domains of the protein, thereby providing exquisite substrate specificity. Disruption of any of the direct substrate-protein residue interactions leads to significant or complete loss of phosphatase activity. Notably, the Tps2PD-BeF3-trehalose complex structure captures an aspartyl-BeF3 covalent adduct, which closely mimics the proposed aspartyl-phosphate intermediate of the phosphatase catalytic cycle. Structures of substrate-free Tps2PD reveal an "open" conformation whereby the cap and core domains separate and visualize the striking conformational changes effected by substrate binding and product release and the role of two hinge regions centered at approximately residues 102-103 and 184-188. Significantly, tps2Δ, tps2NTDΔ, and tps2D705N strains are unable to grow at elevated temperatures. Combined, these studies provide a deeper understanding of the substrate recognition and catalytic mechanism of Tps2 and provide a structural basis for the future design of novel antifungal compounds against a target found in three major fungal pathogens.
Keywords: HASDF phosphatase; antifungal inhibitors; pathogenic fungi; trehalose-6-phosphate phosphatase; trehalose-6-phosphate specificity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
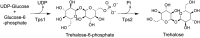


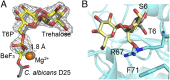

References
-
- Brown GD, et al. Hidden killers: Human fungal infections. Sci Transl Med. 2012;4(165):165rv13. - PubMed
-
- Cowen LE. The evolution of fungal drug resistance: Modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6(3):187–198. - PubMed
-
- Spaltmann F, Blunck M, Ziegelbauer K. Computer-aided target selection-prioritizing targets for antifungal drug discovery. Drug Discov Today. 1999;4(1):17–26. - PubMed
-
- Gancedo C, Flores CL. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res. 2004;4(4-5):351–359. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

