Identification of shared TCR sequences from T cells in human breast cancer using emulsion RT-PCR
- PMID: 27307436
- PMCID: PMC4961128
- DOI: 10.1073/pnas.1606994113
Identification of shared TCR sequences from T cells in human breast cancer using emulsion RT-PCR
Abstract
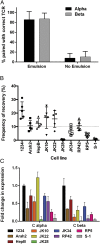
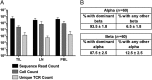
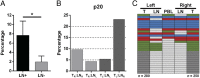
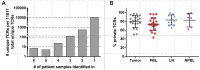
Infiltration of T cells in breast tumors correlates with improved survival of patients with breast cancer, despite relatively few mutations in these tumors. To determine if T-cell specificity can be harnessed to augment immunotherapies of breast cancer, we sought to identify the alpha-beta paired T-cell receptors (TCRs) of tumor-infiltrating lymphocytes shared between multiple patients. Because TCRs function as heterodimeric proteins, we used an emulsion-based RT-PCR assay to link and amplify TCR pairs. Using this assay on engineered T-cell hybridomas, we observed ∼85% accurate pairing fidelity, although TCR recovery frequency varied. When we applied this technique to patient samples, we found that for any given TCR pair, the dominant alpha- or beta-binding partner comprised ∼90% of the total binding partners. Analysis of TCR sequences from primary tumors showed about fourfold more overlap in tumor-involved relative to tumor-free sentinel lymph nodes. Additionally, comparison of sequences from both tumors of a patient with bilateral breast cancer showed 10% overlap. Finally, we identified a panel of unique TCRs shared between patients' tumors and peripheral blood that were not found in the peripheral blood of controls. These TCRs encoded a range of V, J, and complementarity determining region 3 (CDR3) sequences on the alpha-chain, and displayed restricted V-beta use. The nucleotides encoding these shared TCR CDR3s varied, suggesting immune selection of this response. Harnessing these T cells may provide practical strategies to improve the shared antigen-specific response to breast cancer.
Keywords: T-cell receptors; T-cell repertoire profiling; breast cancer; emulsion RT-PCR; high-throughput sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Harnessing shared antigens and T-cell receptors in cancer: Opportunities and challenges.Proc Natl Acad Sci U S A. 2016 Jul 19;113(29):7944-5. doi: 10.1073/pnas.1608860113. Epub 2016 Jul 7. Proc Natl Acad Sci U S A. 2016. PMID: 27410048 Free PMC article. No abstract available.
References
-
- Loi S, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31(7):860–867. - PubMed
-
- Ali HR, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25(8):1536–1543. - PubMed
-
- Lee AH, et al. Different patterns of inflammation and prognosis in invasive carcinoma of the breast. Histopathology. 2006;48(6):692–701. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

