Latency-Associated Nuclear Antigen E3 Ubiquitin Ligase Activity Impacts Gammaherpesvirus-Driven Germinal Center B Cell Proliferation
- PMID: 27307564
- PMCID: PMC4988161
- DOI: 10.1128/JVI.00813-16
Latency-Associated Nuclear Antigen E3 Ubiquitin Ligase Activity Impacts Gammaherpesvirus-Driven Germinal Center B Cell Proliferation
Abstract
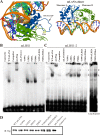
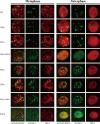
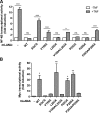
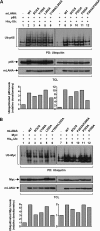
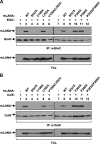
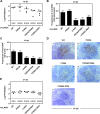
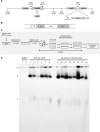
Viruses have evolved mechanisms to hijack components of cellular E3 ubiquitin ligases, thus modulating the ubiquitination pathway. However, the biological relevance of such mechanisms for viral pathogenesis in vivo remains largely unknown. Here, we utilized murid herpesvirus 4 (MuHV-4) infection of mice as a model system to address the role of MuHV-4 latency-associated nuclear antigen (mLANA) E3 ligase activity in gammaherpesvirus latent infection. We show that specific mutations in the mLANA SOCS box (V199A, V199A/L202A, or P203A/P206A) disrupted mLANA's ability to recruit Elongin C and Cullin 5, thereby impairing the formation of the Elongin BC/Cullin 5/SOCS (EC5S(mLANA)) complex and mLANA's E3 ligase activity on host NF-κB and Myc. Although these mutations resulted in considerably reduced mLANA binding to viral terminal repeat DNA as assessed by electrophoretic mobility shift assay (EMSA), the mutations did not disrupt mLANA's ability to mediate episome persistence. In vivo, MuHV-4 recombinant viruses bearing these mLANA SOCS box mutations exhibited a deficit in latency amplification in germinal center (GC) B cells. These findings demonstrate that the E3 ligase activity of mLANA contributes to gammaherpesvirus-driven GC B cell proliferation. Hence, pharmacological inhibition of viral E3 ligase activity through targeting SOCS box motifs is a putative strategy to control gammaherpesvirus-driven lymphoproliferation and associated disease.
Importance: The gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) cause lifelong persistent infection and play causative roles in several human malignancies. Colonization of B cells is crucial for virus persistence, and access to the B cell compartment is gained by virus-driven proliferation in germinal center (GC) B cells. Infection of B cells is predominantly latent, with the viral genome persisting as a multicopy episome and expressing only a small subset of viral genes. Here, we focused on latency-associated nuclear antigen (mLANA) encoded by murid herpesvirus-4 (MuHV-4), which exhibits homology in sequence, structure, and function to KSHV LANA (kLANA), thereby allowing the study of LANA-mediated pathogenesis in mice. Our experiments show that mLANA's E3 ubiquitin ligase activity is necessary for efficient expansion of latency in GC B cells, suggesting that the development of pharmacological inhibitors of LANA E3 ubiquitin ligase activity may allow strategies to interfere with gammaherpesvirus-driven lymphoproliferation and associated disease.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Cross-species conservation of episome maintenance provides a basis for in vivo investigation of Kaposi's sarcoma herpesvirus LANA.PLoS Pathog. 2017 Sep 14;13(9):e1006555. doi: 10.1371/journal.ppat.1006555. eCollection 2017 Sep. PLoS Pathog. 2017. PMID: 28910389 Free PMC article.
-
In Vivo Persistence of Chimeric Virus after Substitution of the Kaposi's Sarcoma-Associated Herpesvirus LANA DNA Binding Domain with That of Murid Herpesvirus 4.J Virol. 2018 Oct 12;92(21):e01251-18. doi: 10.1128/JVI.01251-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111565 Free PMC article.
-
Murine Gammaherpesvirus 68 Expressing Kaposi Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen (LANA) Reveals both Functional Conservation and Divergence in LANA Homologs.J Virol. 2017 Sep 12;91(19):e00992-17. doi: 10.1128/JVI.00992-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28747501 Free PMC article.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
-
E3 Ubiquitin Ligases in Gammaherpesviruses and HIV: A Review of Virus Adaptation and Exploitation.Viruses. 2023 Sep 15;15(9):1935. doi: 10.3390/v15091935. Viruses. 2023. PMID: 37766341 Free PMC article. Review.
Cited by
-
Conquering the Host: Determinants of Pathogenesis Learned from Murine Gammaherpesvirus 68.Annu Rev Virol. 2021 Sep 29;8(1):349-371. doi: 10.1146/annurev-virology-011921-082615. Annu Rev Virol. 2021. PMID: 34586873 Free PMC article.
-
Dangerous Liaisons: Gammaherpesvirus Subversion of the Immunoglobulin Repertoire.Viruses. 2020 Jul 23;12(8):788. doi: 10.3390/v12080788. Viruses. 2020. PMID: 32717815 Free PMC article. Review.
-
Cross-species conservation of episome maintenance provides a basis for in vivo investigation of Kaposi's sarcoma herpesvirus LANA.PLoS Pathog. 2017 Sep 14;13(9):e1006555. doi: 10.1371/journal.ppat.1006555. eCollection 2017 Sep. PLoS Pathog. 2017. PMID: 28910389 Free PMC article.
-
B cell expression of E3 ubiquitin ligase Cul4b promotes chronic gammaherpesvirus infection in vivo.J Virol. 2023 Dec 21;97(12):e0100823. doi: 10.1128/jvi.01008-23. Epub 2023 Nov 14. J Virol. 2023. PMID: 37962378 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

