The N-terminal tropomyosin- and actin-binding sites are important for leiomodin 2's function
- PMID: 27307584
- PMCID: PMC4985258
- DOI: 10.1091/mbc.E16-03-0200
The N-terminal tropomyosin- and actin-binding sites are important for leiomodin 2's function
Abstract
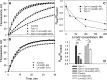
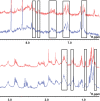
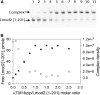
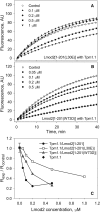
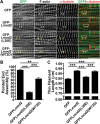
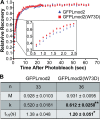
Leiomodin is a potent actin nucleator related to tropomodulin, a capping protein localized at the pointed end of the thin filaments. Mutations in leiomodin-3 are associated with lethal nemaline myopathy in humans, and leiomodin-2-knockout mice present with dilated cardiomyopathy. The arrangement of the N-terminal actin- and tropomyosin-binding sites in leiomodin is contradictory and functionally not well understood. Using one-dimensional nuclear magnetic resonance and the pointed-end actin polymerization assay, we find that leiomodin-2, a major cardiac isoform, has an N-terminal actin-binding site located within residues 43-90. Moreover, for the first time, we obtain evidence that there are additional interactions with actin within residues 124-201. Here we establish that leiomodin interacts with only one tropomyosin molecule, and this is the only site of interaction between leiomodin and tropomyosin. Introduction of mutations in both actin- and tropomyosin-binding sites of leiomodin affected its localization at the pointed ends of the thin filaments in cardiomyocytes. On the basis of our new findings, we propose a model in which leiomodin regulates actin poly-merization dynamics in myocytes by acting as a leaky cap at thin filament pointed ends.
© 2016 Ly, Moroz, et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures


Similar articles
-
Leiomodin creates a leaky cap at the pointed end of actin-thin filaments.PLoS Biol. 2020 Sep 8;18(9):e3000848. doi: 10.1371/journal.pbio.3000848. eCollection 2020 Sep. PLoS Biol. 2020. Update in: PLoS Biol. 2025 Jan 30;23(1):e3003027. doi: 10.1371/journal.pbio.3003027. PMID: 32898131 Free PMC article. Updated.
-
Leiomodin is an actin filament nucleator in muscle cells.Science. 2008 Apr 11;320(5873):239-43. doi: 10.1126/science.1155313. Science. 2008. PMID: 18403713 Free PMC article.
-
Prediction and biological significance of small changes in binding of leiomodin to tropomyosin.J Gen Physiol. 2025 Jul 7;157(4):e202413641. doi: 10.1085/jgp.202413641. Epub 2025 Apr 24. J Gen Physiol. 2025. PMID: 40272387
-
Capping actin filament growth: tropomodulin in muscle and nonmuscle cells.Soc Gen Physiol Ser. 1997;52:79-89. Soc Gen Physiol Ser. 1997. PMID: 9210222 Review.
-
The role of leiomodin in actin dynamics: a new road or a secret gate.FEBS J. 2022 Oct;289(20):6119-6131. doi: 10.1111/febs.16128. Epub 2021 Jul 26. FEBS J. 2022. PMID: 34273242 Free PMC article. Review.
Cited by
-
Characterizing interaction forces between actin and proteins of the tropomodulin family reveals the presence of the N-terminal actin-binding site in leiomodin.Arch Biochem Biophys. 2018 Jan 15;638:18-26. doi: 10.1016/j.abb.2017.12.005. Epub 2017 Dec 6. Arch Biochem Biophys. 2018. PMID: 29223925 Free PMC article.
-
The Mechanisms of Thin Filament Assembly and Length Regulation in Muscles.Int J Mol Sci. 2022 May 10;23(10):5306. doi: 10.3390/ijms23105306. Int J Mol Sci. 2022. PMID: 35628117 Free PMC article. Review.
-
Cardiac leiomodin2 binds to the sides of actin filaments and regulates the ATPase activity of myosin.PLoS One. 2017 Oct 12;12(10):e0186288. doi: 10.1371/journal.pone.0186288. eCollection 2017. PLoS One. 2017. PMID: 29023566 Free PMC article.
-
Tropomodulins and Leiomodins: Actin Pointed End Caps and Nucleators in Muscles.Biophys J. 2017 May 9;112(9):1742-1760. doi: 10.1016/j.bpj.2017.03.034. Biophys J. 2017. PMID: 28494946 Free PMC article. Review.
-
The cardiomyopathy-associated K15N mutation in tropomyosin alters actin filament pointed end dynamics.Arch Biochem Biophys. 2017 Sep 15;630:18-26. doi: 10.1016/j.abb.2017.07.006. Epub 2017 Jul 18. Arch Biochem Biophys. 2017. PMID: 28732641 Free PMC article.
References
-
- Brand NJ, Lara-Pezzi E, Rosenthal N, Barton PJ. Analysis of cardiac myocyte biology in transgenic mice: a protocol for preparation of neonatal mouse cardiac myocyte cultures. Methods Mol Biol. 2010;633:113–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

