BMI-1 Targeting Interferes with Patient-Derived Tumor-Initiating Cell Survival and Tumor Growth in Prostate Cancer
- PMID: 27307599
- PMCID: PMC5159329
- DOI: 10.1158/1078-0432.CCR-15-3107
BMI-1 Targeting Interferes with Patient-Derived Tumor-Initiating Cell Survival and Tumor Growth in Prostate Cancer
Abstract
Purpose: Current prostate cancer management calls for identifying novel and more effective therapies. Self-renewing tumor-initiating cells (TICs) hold intrinsic therapy resistance and account for tumor relapse and progression. As BMI-1 regulates stem cell self-renewal, impairing BMI-1 function for TIC-tailored therapies appears to be a promising approach.
Experimental design: We have previously developed a combined immunophenotypic and time-of-adherence assay to identify CD49bhiCD29hiCD44hi cells as human prostate TICs. We utilized this assay with patient-derived prostate cancer cells and xenograft models to characterize the effects of pharmacologic inhibitors of BMI-1.
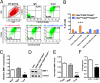
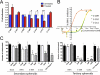
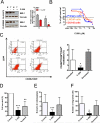
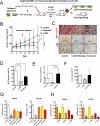
Results: We demonstrate that in cell lines and patient-derived TICs, BMI-1 expression is upregulated and associated with stem cell-like traits. From a screened library, we identified a number of post-transcriptional small molecules that target BMI-1 in prostate TICs. Pharmacologic inhibition of BMI-1 in patient-derived cells significantly decreased colony formation in vitro and attenuated tumor initiation in vivo, thereby functionally diminishing the frequency of TICs, particularly in cells resistant to proliferation- and androgen receptor-directed therapies, without toxic effects on normal tissues.
Conclusions: Our data offer a paradigm for targeting TICs and support the development of BMI-1-targeting therapy for a more effective prostate cancer treatment. Clin Cancer Res; 22(24); 6176-91. ©2016 AACR.
©2016 American Association for Cancer Research.
Figures


References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. - PubMed
-
- Attard G, Parker C, Eeles RA, Schroder F, Tomlins SA, Tannock I, et al. Prostate cancer. Lancet. 2016;387:70–82. - PubMed
-
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51. - PubMed
-
- Toivanen R, Berman DM, Wang H, Pedersen J, Frydenberg M, Meeker AK, et al. Brief report: a bioassay to identify primary human prostate cancer repopulating cells. Stem Cells. 2011;29:1310–4. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

