Read-based phasing of related individuals
- PMID: 27307622
- PMCID: PMC4908360
- DOI: 10.1093/bioinformatics/btw276
Read-based phasing of related individuals
Abstract
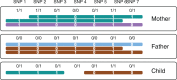
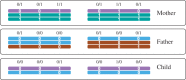
Motivation: Read-based phasing deduces the haplotypes of an individual from sequencing reads that cover multiple variants, while genetic phasing takes only genotypes as input and applies the rules of Mendelian inheritance to infer haplotypes within a pedigree of individuals. Combining both into an approach that uses these two independent sources of information-reads and pedigree-has the potential to deliver results better than each individually.
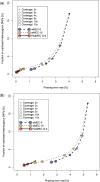
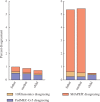
Results: We provide a theoretical framework combining read-based phasing with genetic haplotyping, and describe a fixed-parameter algorithm and its implementation for finding an optimal solution. We show that leveraging reads of related individuals jointly in this way yields more phased variants and at a higher accuracy than when phased separately, both in simulated and real data. Coverages as low as 2× for each member of a trio yield haplotypes that are as accurate as when analyzed separately at 15× coverage per individual.
Availability and implementation: https://bitbucket.org/whatshap/whatshap
Contact: t.marschall@mpi-inf.mpg.de.
© The Author 2016. Published by Oxford University Press.
Figures
References
-
- Chen Z.Z. et al. (2013b) Exact algorithms for haplotype assembly from whole-genome sequence data. Bioinformatics, 29, 1938–1945. - PubMed
-
- Cilibrasi R. et al. (2007) The complexity of the single individual SNP haplotyping problem. Algorithmica, 49, 13–36.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

