Fast metabolite identification with Input Output Kernel Regression
- PMID: 27307628
- PMCID: PMC4908330
- DOI: 10.1093/bioinformatics/btw246
Fast metabolite identification with Input Output Kernel Regression
Abstract
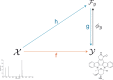
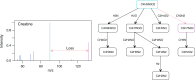
Motivation: An important problematic of metabolomics is to identify metabolites using tandem mass spectrometry data. Machine learning methods have been proposed recently to solve this problem by predicting molecular fingerprint vectors and matching these fingerprints against existing molecular structure databases. In this work we propose to address the metabolite identification problem using a structured output prediction approach. This type of approach is not limited to vector output space and can handle structured output space such as the molecule space.
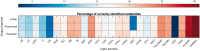
Results: We use the Input Output Kernel Regression method to learn the mapping between tandem mass spectra and molecular structures. The principle of this method is to encode the similarities in the input (spectra) space and the similarities in the output (molecule) space using two kernel functions. This method approximates the spectra-molecule mapping in two phases. The first phase corresponds to a regression problem from the input space to the feature space associated to the output kernel. The second phase is a preimage problem, consisting in mapping back the predicted output feature vectors to the molecule space. We show that our approach achieves state-of-the-art accuracy in metabolite identification. Moreover, our method has the advantage of decreasing the running times for the training step and the test step by several orders of magnitude over the preceding methods.
Contact: celine.brouard@aalto.fi
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press.
Figures




References
-
- Allen F. et al. (2015) Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics, 11, 98–110.
-
- Böcker S., Rasche F. (2008) Towards de novo identification of metabolites by analyzing tandem mass spectra. Bioinfomatics, 24, i49–i55. - PubMed
-
- Bolton E. et al. (2008). PubChem: Integrated platform of small molecules and biological activities. Chapter 12 in Annual Reports in Computational Chemistry, vol. 4, pp. 217–241.
-
- Brouard C. et al. (2011). Semi-supervised penalized output kernel regression for link prediction. In Proceedings of the 28th International Conference on Machine Learning, pp. 593–600. Bellevue, Washington, USA.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

