What time is it? Deep learning approaches for circadian rhythms
- PMID: 27307647
- PMCID: PMC4908327
- DOI: 10.1093/bioinformatics/btw243
What time is it? Deep learning approaches for circadian rhythms
Erratum in
-
What time is it? Deep learning approaches for circadian rhythms.Bioinformatics. 2016 Oct 1;32(19):3051. doi: 10.1093/bioinformatics/btw504. Epub 2016 Aug 19. Bioinformatics. 2016. PMID: 27542773 Free PMC article. No abstract available.
Abstract
Motivation: Circadian rhythms date back to the origins of life, are found in virtually every species and every cell, and play fundamental roles in functions ranging from metabolism to cognition. Modern high-throughput technologies allow the measurement of concentrations of transcripts, metabolites and other species along the circadian cycle creating novel computational challenges and opportunities, including the problems of inferring whether a given species oscillate in circadian fashion or not, and inferring the time at which a set of measurements was taken.
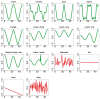
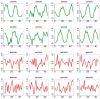
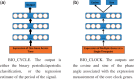
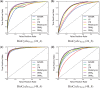
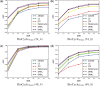
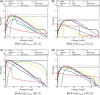
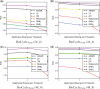
Results: We first curate several large synthetic and biological time series datasets containing labels for both periodic and aperiodic signals. We then use deep learning methods to develop and train BIO_CYCLE, a system to robustly estimate which signals are periodic in high-throughput circadian experiments, producing estimates of amplitudes, periods, phases, as well as several statistical significance measures. Using the curated data, BIO_CYCLE is compared to other approaches and shown to achieve state-of-the-art performance across multiple metrics. We then use deep learning methods to develop and train BIO_CLOCK to robustly estimate the time at which a particular single-time-point transcriptomic experiment was carried. In most cases, BIO_CLOCK can reliably predict time, within approximately 1 h, using the expression levels of only a small number of core clock genes. BIO_CLOCK is shown to work reasonably well across tissue types, and often with only small degradation across conditions. BIO_CLOCK is used to annotate most mouse experiments found in the GEO database with an inferred time stamp.
Availability and implementation: All data and software are publicly available on the CircadiOmics web portal: circadiomics.igb.uci.edu/
Contacts: fagostin@uci.edu or pfbaldi@uci.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press.
Figures


References
-
- Allison D.B. et al. (2002) A mixture model approach for the analysis of microarray gene expression data. Comput. Stat. Data Anal., 39, 1–20.
-
- Antunes L.C. et al. (2010) Obesity and shift work: chronobiological aspects. Nutr. Res. Rev., 23, 155–168. - PubMed
-
- Baldi P. (2012) Autoencoders, Unsupervised Learning, and Deep Architectures. J. Mach. Learn. Res., 27, 37–50 (Proceedings of 2011 ICML Workshop on Unsupervised and Transfer Learning).

