Role of the double-strand break repair pathway in the maintenance of genomic stability
- PMID: 27308383
- PMCID: PMC4905226
- DOI: 10.4161/23723548.2014.968020
Role of the double-strand break repair pathway in the maintenance of genomic stability
Abstract
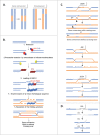
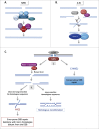
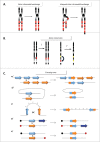
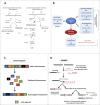
DNA double-strand breaks (DSBs) are highly lethal lesions that jeopardize genome integrity. However, DSBs are also used to generate diversity during the physiological processes of meiosis or establishment of the immune repertoire. Therefore, DSB repair must be tightly controlled. Two main strategies are used to repair DSBs: homologous recombination (HR) and non-homologous end joining (NHEJ). HR is generally considered to be error-free, whereas NHEJ is considered to be error-prone. However, recent data challenge these assertions. Here, we present the molecular mechanisms involved in HR and NHEJ and the recently described alternative end-joining mechanism, which is exclusively mutagenic. Whereas NHEJ is not intrinsically error-prone but adaptable, HR has the intrinsic ability to modify the DNA sequence. Importantly, in both cases the initial structure of the DNA impacts the outcome. Finally, the consequences and applications of these repair mechanisms are discussed. Both HR and NHEJ are double-edged swords, essential for maintenance of genome stability and diversity but also able to generate genome instability.
Keywords: DNA repair; double-strand break repair; genome instability; genome rearrangements; homologous recombination; ionizing radiation; mutagenesis; non-homologous end joining; telomeres.
Figures
References
-
- Haber JE. Genome stability. DNA Repair Recombination. Talor and Francis Group: New York; 2014.
-
- Magdalou I, Lopez BS, Pasero P, Lambert SA. The causes of replication stress and their consequences on genome stability and cell fate. Semin Cell Dev Biol 2014; 30C:154-64; http://dx.doi.org/10.1016/j.semcdb.2014.04.035 - DOI - PubMed
-
- Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet 2014; 10:e1004086; PMID: 24453986; http://dx.doi.org/10.1371/journal.pgen.1004086 - DOI - PMC - PubMed
-
- Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 2009; 16:819-24; PMID: 19633668; http://dx.doi.org/10.1038/nsmb.1641 - DOI - PubMed
-
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 2001; 15:3237-42; PMID: 11751629; http://dx.doi.org/10.1101/gad.946401 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
