Design of a hyperstable 60-subunit protein dodecahedron. [corrected]
- PMID: 27309817
- PMCID: PMC4945409
- DOI: 10.1038/nature18010
Design of a hyperstable 60-subunit protein dodecahedron. [corrected]
Erratum in
-
Corrigendum: Design of a hyperstable 60-subunit protein icosahedron.Nature. 2016 Dec 1;540(7631):150. doi: 10.1038/nature20108. Epub 2016 Oct 19. Nature. 2016. PMID: 27760110 No abstract available.
Abstract
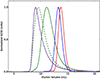
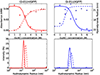
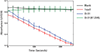
The dodecahedron [corrected] is the largest of the Platonic solids, and icosahedral protein structures are widely used in biological systems for packaging and transport. There has been considerable interest in repurposing such structures for applications ranging from targeted delivery to multivalent immunogen presentation. The ability to design proteins that self-assemble into precisely specified, highly ordered icosahedral structures would open the door to a new generation of protein containers with properties custom-tailored to specific applications. Here we describe the computational design of a 25-nanometre icosahedral nanocage that self-assembles from trimeric protein building blocks. The designed protein was produced in Escherichia coli, and found by electron microscopy to assemble into a homogenous population of icosahedral particles nearly identical to the design model. The particles are stable in 6.7 molar guanidine hydrochloride at up to 80 degrees Celsius, and undergo extremely abrupt, but reversible, disassembly between 2 molar and 2.25 molar guanidinium thiocyanate. The dodecahedron [corrected] is robust to genetic fusions: one or two copies of green fluorescent protein (GFP) can be fused to each of the 60 subunits to create highly fluorescent ‘standard candles’ for use in light microscopy, and a designed protein pentamer can be placed in the centre of each of the 20 pentameric faces to modulate the size of the entrance/exit channels of the cage. Such robust and customizable nanocages should have considerable utility in targeted drug delivery, vaccine design and synthetic biology.
Figures






References
-
- Ritsert K, et al. Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: crystal structure analysis of reconstituted, icosahedral beta-subunit capsids with bound substrate analogue inhibitor at 2.4 A resolution. J. Mol. Biol. 1995;253:151–167. - PubMed
-
- Howorka S. Rationally engineering natural protein assemblies in nanobiotechnology. Curr. Opin. Biotechnol. 2011;22:485–491. - PubMed
-
- Roldão A, Mellado MCM, Castilho LR, Carrondo MJT, Alves PM. Virus-like particles in vaccine development. Expert Rev. Vaccines. 2010;9:1149–1176. - PubMed
-
- Effio CL, Hubbuch J. Next generation vaccines and vectors: Designing downstream processes for recombinant protein-based virus-like particles. Biotechnol. J. 2015;10:715–727. - PubMed
References for online-only Methods
-
- Zhou Z, et al. Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS Chem. Biol. 2007;2:337–346. - PubMed
-
- Baalousha M, Lead JR. Nanoparticle dispersity in toxicology. Nat. Nanotechnol. 2013;8:308–309. - PubMed
-
- Zulauf M, D’Arcy A. Light scattering of proteins as a criterion for crystallization. J. Cryst. Growth. 1992;122:102–106.
-
- Nannenga BL, Iadanza MG, Vollmar BS, Gonen T. In: Current Protocols in Protein Science. Coligan JE, Dunn BM, Speicher DW, Wingfield PT, editors. John Wiley & Sons Inc.; 2013.
-
- Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. - PubMed

