The eukaryotic CMG helicase pumpjack and integration into the replisome
- PMID: 27310307
- PMCID: PMC4916876
- DOI: 10.1080/19491034.2016.1174800
The eukaryotic CMG helicase pumpjack and integration into the replisome
Abstract
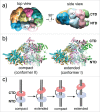
The eukaryotic replisome is α multiprotein machine that contains DNA polymerases, sliding clamps, helicase, and primase along with several factors that participate in cell cycle and checkpoint control. The detailed structure of the 11-subunit CMG helicase (Cdc45/Mcm2-7/GINS) has been solved recently by cryoEM single-particle 3D reconstruction and reveals pumpjack motions that imply an unexpected mechanism of DNA translocation. CMG is also the organizing center of the replisome. Recent in vitro reconstitution of leading and lagging strand DNA synthesis has enabled structural analysis of the replisome. By building the replisome in stages from pure proteins, single-particle EM studies have identified the overall architecture of the eukaryotic replisome. Suprisingly leading and lagging strand polymerases bind to opposite faces of the CMG helicase, unlike the long-held view that DNA polymerases are located in back of the helicase to act on the unwound strands.
Keywords: 3D reconstruction; CMG; DNA helicase; DNA polymerase; cryoelectron microscopy; primase; replication fork; replisome.
Figures


References
-
- O'Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol 2013; 5:a010108; PMID:23818497; http://dx.doi.org/10.1101/cshperspect.a010108 - DOI - PMC - PubMed
-
- MacNeill S. The Eukaryotic Replisome: A Guide to Protein Structure and Function. Springer; Netherlands, 2012
-
- Diffley JF. On the road to replication. EMBO Mol Med 2016; 8:77-9; PMID:26787652; http://dx.doi.org/10.15252/emmm.201505965 - DOI - PMC - PubMed
-
- Sun J, Fernandez-Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, Speck C, Li H. Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function. Genes Dev 2014; 28:2291-303; PMID:25319829; http://dx.doi.org/10.1101/gad.242313.114 - DOI - PMC - PubMed
-
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A 2009; 106:20240-5; PMID:19910535; http://dx.doi.org/10.1073/pnas.0911500106 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
