Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection
- PMID: 27310713
- PMCID: PMC4911051
- DOI: 10.1371/journal.pone.0157368
Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection
Abstract
Triple-negative breast cancer (TNBC) is a heterogeneous disease that can be classified into distinct molecular subtypes by gene expression profiling. Considered a difficult-to-treat cancer, a fraction of TNBC patients benefit significantly from neoadjuvant chemotherapy and have far better overall survival. Outside of BRCA1/2 mutation status, biomarkers do not exist to identify patients most likely to respond to current chemotherapy; and, to date, no FDA-approved targeted therapies are available for TNBC patients. Previously, we developed an approach to identify six molecular subtypes TNBC (TNBCtype), with each subtype displaying unique ontologies and differential response to standard-of-care chemotherapy. Given the complexity of the varying histological landscape of tumor specimens, we used histopathological quantification and laser-capture microdissection to determine that transcripts in the previously described immunomodulatory (IM) and mesenchymal stem-like (MSL) subtypes were contributed from infiltrating lymphocytes and tumor-associated stromal cells, respectively. Therefore, we refined TNBC molecular subtypes from six (TNBCtype) into four (TNBCtype-4) tumor-specific subtypes (BL1, BL2, M and LAR) and demonstrate differences in diagnosis age, grade, local and distant disease progression and histopathology. Using five publicly available, neoadjuvant chemotherapy breast cancer gene expression datasets, we retrospectively evaluated chemotherapy response of over 300 TNBC patients from pretreatment biopsies subtyped using either the intrinsic (PAM50) or TNBCtype approaches. Combined analysis of TNBC patients demonstrated that TNBC subtypes significantly differ in response to similar neoadjuvant chemotherapy with 41% of BL1 patients achieving a pathological complete response compared to 18% for BL2 and 29% for LAR with 95% confidence intervals (CIs; [33, 51], [9, 28], [17, 41], respectively). Collectively, we provide pre-clinical data that could inform clinical trials designed to test the hypothesis that improved outcomes can be achieved for TNBC patients, if selection and combination of existing chemotherapies is directed by knowledge of molecular TNBC subtypes.
Conflict of interest statement
Figures
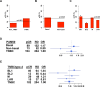
References
-
- Esserman LJ, Berry DA, Cheang MCU, Yau C, Perou CM, Carey L, et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res Treat. 2012;132: 1049–1062. 10.1007/s10549-011-1895-2 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

