Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery
- PMID: 27311775
- PMCID: PMC5423194
- DOI: 10.1016/j.molonc.2016.05.009
Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery
Abstract
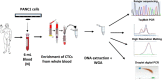
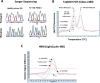
In colorectal cancer (CRC), KRAS mutations are a strong negative predictor for treatment with the EGFR-targeted antibodies cetuximab and panitumumab. Since it can be difficult to obtain appropriate tumor tissues for KRAS genotyping, alternative methods are required. Circulating tumor cells (CTCs) are believed to be representative of the tumor in real time. In this study we explored the capacity of a size-based device for capturing CTCs coupled with a multiplex KRAS screening assay using droplet digital PCR (ddPCR). We showed that it is possible to detect a mutant ratio of 0.05% and less than one KRAS mutant cell per mL total blood with ddPCR compared to about 0.5% and 50-75 cells for TaqMeltPCR and HRM. Next, CTCs were isolated from the blood of 35 patients with CRC at various stage of the disease. KRAS genotyping was successful for 86% (30/35) of samples with a KRAS codon 12/13 mutant ratio of 57% (17/30). In contrast, only one patient was identified as KRAS mutant when size-based isolation was combined with HRM or TaqMeltPCR. KRAS status was then determined for the 26 available formalin-fixed paraffin-embedded tumors using standard procedures. The concordance between the CTCs and the corresponding tumor tissues was 77% with a sensitivity of 83%. Taken together, the data presented here suggest that is feasible to detect KRAS mutations in CTCs from blood samples of CRC patients which are predictive for those found in the tumor. The minimal invasive nature of this procedure in combination with the high sensitivity of ddPCR might provide in the future an opportunity to monitor patients throughout the course of disease on multiple levels including early detection, prognosis, treatment and relapse as well as to obtain mechanistic insight with respect to tumor invasion and metastasis.
Keywords: Circulating tumor cells; Colorectal cancer; Digital PCR; KRAS; Liquid biopsy.
Copyright © 2016 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Alix-Panabieres, C. , Pantel, K. , 2014. Technologies for detection of circulating tumor cells: facts and vision. Lab Chip 14, 57–62. - PubMed
-
- Amado, R.G. , Wolf, M. , Peeters, M. , Van Cutsem, E. , Siena, S. , Freeman, D.J. , Juan, T. , Sikorski, R. , Suggs, S. , Radinsky, R. , Patterson, S.D. , Chang, D.D. , 2008. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 26, 1626–1634. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

