Microbial interactions lead to rapid micro-scale successions on model marine particles
- PMID: 27311813
- PMCID: PMC4915023
- DOI: 10.1038/ncomms11965
Microbial interactions lead to rapid micro-scale successions on model marine particles
Abstract
In the ocean, organic particles harbour diverse bacterial communities, which collectively digest and recycle essential nutrients. Traits like motility and exo-enzyme production allow individual taxa to colonize and exploit particle resources, but it remains unclear how community dynamics emerge from these individual traits. Here we track the taxon and trait dynamics of bacteria attached to model marine particles and demonstrate that particle-attached communities undergo rapid, reproducible successions driven by ecological interactions. Motile, particle-degrading taxa are selected for during early successional stages. However, this selective pressure is later relaxed when secondary consumers invade, which are unable to use the particle resource but, instead, rely on carbon from primary degraders. This creates a trophic chain that shifts community metabolism away from the particle substrate. These results suggest that primary successions may shape particle-attached bacterial communities in the ocean and that rapid community-wide metabolic shifts could limit rates of marine particle degradation.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
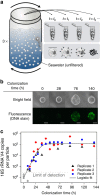
 ,
,  ,
,  ). Symbols in grey (
). Symbols in grey ( ,
,  ,
,  ) indicate measurements below the limit of detection of the assay. The grey line (-) indicates the fit to a logistic growth model.
) indicate measurements below the limit of detection of the assay. The grey line (-) indicates the fit to a logistic growth model.
 ,
,  ,
,  ). Grey lines indicate the median trajectories. (d) Histogram of cross-replicate correlations for individual taxa (Methods). (e) Shannon diversity (
). Grey lines indicate the median trajectories. (d) Histogram of cross-replicate correlations for individual taxa (Methods). (e) Shannon diversity ( ) over time for the three colonization replicates (
) over time for the three colonization replicates ( ,
,  ,
,  ). Samples for which sequencing coverage was insufficient for the Shannon diversity to saturate have been omitted. The solid grey line (-) indicates the initial Shannon diversity of the seawater.
). Samples for which sequencing coverage was insufficient for the Shannon diversity to saturate have been omitted. The solid grey line (-) indicates the initial Shannon diversity of the seawater.
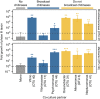
References
-
- Alldredge A. L. & Silver M. W. Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 20, 41–82 (1988).
-
- Simon M., Grossart H. P., Schweitzer B. & Ploug H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211 (2002).
-
- Jiao N. et al. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8, 593–599 (2010). - PubMed
-
- Kirchman D. L. Microbial Ecology of the Oceans John Wiley & Sons (2010).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

