B-cell repertoire dynamics after sequential hepatitis B vaccination and evidence for cross-reactive B-cell activation
- PMID: 27312086
- PMCID: PMC4910312
- DOI: 10.1186/s13073-016-0322-z
B-cell repertoire dynamics after sequential hepatitis B vaccination and evidence for cross-reactive B-cell activation
Erratum in
-
Erratum to: B-cell repertoire dynamics after sequential hepatitis B vaccination and evidence for cross-reactive B-cell activation.Genome Med. 2016 Aug 3;8(1):81. doi: 10.1186/s13073-016-0337-5. Genome Med. 2016. PMID: 27488676 Free PMC article. No abstract available.
Abstract
Background: A diverse B-cell repertoire is essential for recognition and response to infectious and vaccine antigens. High-throughput sequencing of B-cell receptor (BCR) genes can now be used to study the B-cell repertoire at great depth and may shed more light on B-cell responses than conventional immunological methods. Here, we use high-throughput BCR sequencing to provide novel insight into B-cell dynamics following a primary course of hepatitis B vaccination.
Methods: Nine vaccine-naïve participants were administered three doses of hepatitis B vaccine (months 0, 1, and 2 or 7). High-throughput Illumina sequencing of the total BCR repertoire was combined with targeted sequencing of sorted vaccine antigen-enriched B cells to analyze the longitudinal response of both the total and vaccine-specific repertoire after each vaccine. ELISpot was used to determine vaccine-specific cell numbers following each vaccine.
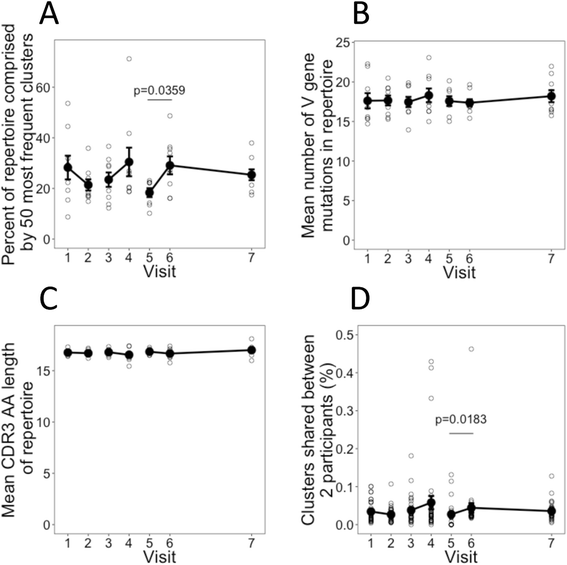
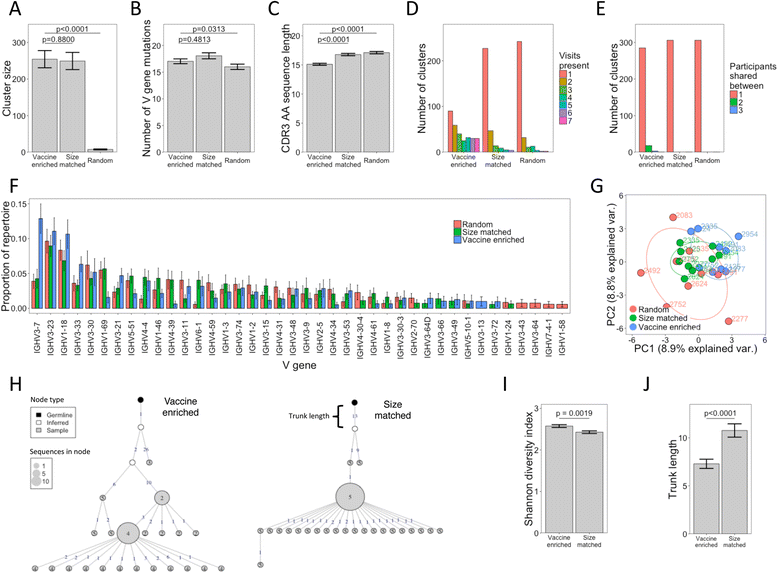
Results: Deconvoluting the vaccine-specific from total BCR repertoire showed that vaccine-specific sequence clusters comprised <0.1 % of total sequence clusters, and had certain stereotypic features. The vaccine-specific BCR sequence clusters were expanded after each of the three vaccine doses, despite no vaccine-specific B cells being detected by ELISpot after the first vaccine dose. These vaccine-specific BCR clusters detected after the first vaccine dose had distinct properties compared to those detected after subsequent doses; they were more mutated, present at low frequency even prior to vaccination, and appeared to be derived from more mature B cells.
Conclusions: These results demonstrate the high-sensitivity of our vaccine-specific BCR analysis approach and suggest an alternative view of the B-cell response to novel antigens. In the response to the first vaccine dose, many vaccine-specific BCR clusters appeared to largely derive from previously activated cross-reactive B cells that have low affinity for the vaccine antigen, and subsequent doses were required to yield higher affinity B cells.
Keywords: Antibody; B-cell repertoire; Hepatitis B; Polyreactive; VDJ; Vaccination.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

