Molecular Landscape of Acquired Resistance to Targeted Therapy Combinations in BRAF-Mutant Colorectal Cancer
- PMID: 27312529
- PMCID: PMC4970882
- DOI: 10.1158/0008-5472.CAN-16-0396
Molecular Landscape of Acquired Resistance to Targeted Therapy Combinations in BRAF-Mutant Colorectal Cancer
Abstract
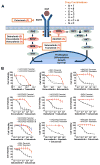
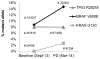
Although recent clinical trials of BRAF inhibitor combinations have demonstrated improved efficacy in BRAF-mutant colorectal cancer, emergence of acquired resistance limits clinical benefit. Here, we undertook a comprehensive effort to define mechanisms underlying drug resistance with the goal of guiding development of therapeutic strategies to overcome this limitation. We generated a broad panel of BRAF-mutant resistant cell line models across seven different clinically relevant drug combinations. Combinatorial drug treatments were able to abrogate ERK1/2 phosphorylation in parental-sensitive cells, but not in their resistant counterparts, indicating that resistant cells escaped drug treatments through one or more mechanisms leading to biochemical reactivation of the MAPK signaling pathway. Genotyping of resistant cells identified gene amplification of EGFR, KRAS, and mutant BRAF, as well as acquired mutations in KRAS, EGFR, and MAP2K1 These mechanisms were clinically relevant, as we identified emergence of a KRAS G12C mutation and increase of mutant BRAF V600E allele frequency in the circulating tumor DNA of a patient at relapse from combined treatment with BRAF and MEK inhibitors. To identify therapeutic combinations capable of overcoming drug resistance, we performed a systematic assessment of candidate therapies across the panel of resistant cell lines. Independent of the molecular alteration acquired upon drug pressure, most resistant cells retained sensitivity to vertical MAPK pathway suppression when combinations of ERK, BRAF, and EGFR inhibitors were applied. These therapeutic combinations represent promising strategies for future clinical trials in BRAF-mutant colorectal cancer. Cancer Res; 76(15); 4504-15. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
Figures




References
-
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. - PubMed
-
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

