Translation dynamics of single mRNAs in live cells and neurons
- PMID: 27313041
- PMCID: PMC4939616
- DOI: 10.1126/science.aaf1084
Translation dynamics of single mRNAs in live cells and neurons
Abstract
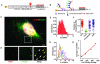
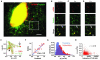
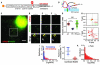
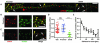
Translation is the fundamental biological process converting mRNA information into proteins. Single-molecule imaging in live cells has illuminated the dynamics of RNA transcription; however, it is not yet applicable to translation. Here, we report single-molecule imaging of nascent peptides (SINAPS) to assess translation in live cells. The approach provides direct readout of initiation, elongation, and location of translation. We show that mRNAs coding for endoplasmic reticulum (ER) proteins are translated when they encounter the ER membrane. Single-molecule fluorescence recovery after photobleaching provides direct measurement of elongation speed (5 amino acids per second). In primary neurons, mRNAs are translated in proximal dendrites but repressed in distal dendrites and display "bursting" translation. This technology provides a tool with which to address the spatiotemporal translation mechanism of single mRNAs in living cells.
Copyright © 2016, American Association for the Advancement of Science.
Figures

Comment in
-
Translation: Live stream: translation at single-mRNA resolution.Nat Rev Genet. 2016 Jun 16;17(7):373. doi: 10.1038/nrg.2016.79. Nat Rev Genet. 2016. PMID: 27306872 No abstract available.
-
PROTEIN TRANSLATION. Seeing translation.Science. 2016 Jun 17;352(6292):1391-2. doi: 10.1126/science.aag1039. Science. 2016. PMID: 27313023 No abstract available.
-
Tracking translation one mRNA at a time.Nat Biotechnol. 2016 Jul 12;34(7):723-4. doi: 10.1038/nbt.3632. Nat Biotechnol. 2016. PMID: 27404883 No abstract available.
References
-
- Schwanhäusser B, et al. Nature. 2011;473:337–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

