Expect the Unexpected Enrichment of "Hidden Proteome" of Seeds and Tubers by Depletion of Storage Proteins
- PMID: 27313590
- PMCID: PMC4887479
- DOI: 10.3389/fpls.2016.00761
Expect the Unexpected Enrichment of "Hidden Proteome" of Seeds and Tubers by Depletion of Storage Proteins
Abstract
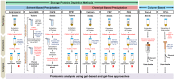
Dynamic resolution of seed and tuber protein samples is highly limited due to the presence of high-abundance storage proteins (SPs). These proteins inevitably obscure the low-abundance proteins (LAPs) impeding their identification and characterization. To facilitate the detection of LAPs, several methods have been developed during the past decade, enriching the proteome with extreme proteins. Most of these methods, if not all, are based on the specific removal of SPs which ultimately magnify the proteome coverage. In this mini-review, we summarize the available methods that have been developed over the years for the enrichment of LAPs either from seeds or tubers. Incorporation of these methods during the protein extraction step will be helpful in understanding the seed/tuber biology in greater detail.
Keywords: depletion methods; hidden proteome; high-abundance proteins; low-abundance proteins; plant; proteome coverage; storage proteins.
Figures
References
-
- Agrawal G. K., Rakwal R. (2012). Seed Development: OMICS Technologies Toward Improvement of Seed Quality and Crop Yield. Berlin: Springer; 10.1007/978-94-007-4749-4 - DOI
-
- Boschetti E., Righetti P. G. (2013). Low-Abundance Proteome Discovery: State of the Art and Protocols. Oxford: Newnes.
-
- Fröhlich A., Gaupels F., Sarioglu H., Holzmeister C., Spannagl M., Durner J., et al. (2012). Looking deep inside: detection of low-abundant proteins in leave extracts of Arabidopsis thaliana and phloem exudates of Cucurbita maxima. Plant Physiol. 159 902–914. 10.1104/pp.112.198077 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous