Identification and Validation of Reference Genes for qRT-PCR Studies of Gene Expression in Dioscorea opposita
- PMID: 27314014
- PMCID: PMC4899605
- DOI: 10.1155/2016/3089584
Identification and Validation of Reference Genes for qRT-PCR Studies of Gene Expression in Dioscorea opposita
Abstract
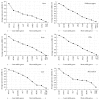
Quantitative real-time polymerase chain reaction (qRT-PCR) is one of the most common methods for gene expression studies. Data normalization based on reference genes is essential for obtaining reliable results for qRT-PCR assays. This study evaluated potential reference genes of Chinese yam (Dioscorea opposita Thunb.), which is an important tuber crop and medicinal plant in East Asia. The expression of ten candidate reference genes across 20 samples from different organs and development stages was assessed. We identified the most stable genes for qRT-PCR studies using combined samples from different organs. Our results also suggest that different suitable reference genes or combinations of reference genes for normalization should be applied according to different organs and developmental stages. To validate the suitability of the reference genes, we evaluated the relative expression of PE2.1 and PE53, which are two genes that may be associated with microtuber formation. Our results provide the foundation for reference gene(s) selection in D. opposita and will contribute toward more accurate gene analysis studies of the genus Dioscorea.
Figures




References
-
- Artico S., Nardeli S. M., Brilhante O., Grossi-de-Sa M. F., Alves-Ferreira M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biology. 2010;10, article 49 doi: 10.1186/1471-2229-10-49. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

