Extended-Spectrum Cephalosporin-Resistant Salmonella enterica serovar Heidelberg Strains, the Netherlands(1)
- PMID: 27314180
- PMCID: PMC4918182
- DOI: 10.3201/eid2207.151377
Extended-Spectrum Cephalosporin-Resistant Salmonella enterica serovar Heidelberg Strains, the Netherlands(1)
Abstract
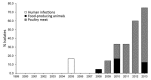
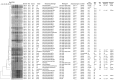
Extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg strains (JF6X01.0022/XbaI.0251, JF6X01.0326/XbaI.1966, JF6X01.0258/XbaI.1968, and JF6X01.0045/XbaI.1970) have been identified in the United States with pulsed-field gel electrophoresis. Our examination of isolates showed introduction of these strains in the Netherlands and highlight the need for active surveillance and intervention strategies by public health organizations.
Keywords: AmpC; IncI1 plasmids; Salmonella enterica serovar Heidelberg; Salmonella infections; antibacterial agents; antimicrobial resistance; bacteria; extended-spectrum cephalosporins; the Netherlands.
Figures
References
-
- European Centre for Disease Prevention and Control. Technical document. EU protocol for harmonised monitoring of antimicrobial resistance in human Salmonella and Campylobacter isolates. Stockholm: The Centre; 2014.
-
- Public Health Agency of Canada. National Enteric Surveillance Program (NESP). Annual summary 2012. Ottawa (Ontario, CA): The Agency; 2014.
-
- Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System: enteric bacteria. 2012 Human isolates final report. Atlanta: The Centers; 2014.
-
- Hoffmann M, Zhao S, Pettengill J, Luo Y, Monday SR, Abbott J, et al. Comparative genomic analysis and virulence differences in closely related Salmonella enterica serotype Heidelberg isolates from humans, retail meats, and animals. Genome Biol Evol. 2014;6:1046–68. 10.1093/gbe/evu079 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

