HCV NS4B targets Scribble for proteasome-mediated degradation to facilitate cell transformation
- PMID: 27315218
- PMCID: PMC7097421
- DOI: 10.1007/s13277-016-5100-4
HCV NS4B targets Scribble for proteasome-mediated degradation to facilitate cell transformation
Abstract
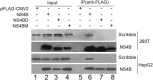
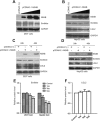
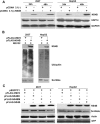
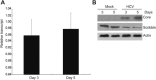
Hepatitis C virus (HCV) nonstructural protein 4B (NS4B) is a multi-transmembrane protein, but little is known about how NS4B contributes to HCV replication and tumorigenesis. Its C-terminal domain (CTD) has been shown to associate with intracellular membrane, and we have previously shown that NS4B CTD contains a class I PDZ-binding motif (PBM). Here, we demonstrated that NS4B PBM interacts with the PDZ-containing tumor suppressor protein, Scribble, using immunofluorescence and co-immunoprecipitation assays, and this interaction requires at least three contiguous PDZ domains of Scribble. In addition, NS4B PBM specifically induced Scribble degradation by activating the proteasome-ubiquitin pathway. Similar Scribble degradation was also observed in HCV-infected cells, suggesting NS4B could work in the context of HCV. Finally, NS4B PBM mutants showed reduced colony formation capacity compared with its wild-type counterpart, indicating that NS4B PBM plays important roles in NS4B-mediated cell transformation. Altogether, we provide a mechanism by which NS4B induces cell transformation through its PBM, which specifically interacts with the PDZ domains of Scribble and targets Scribble for degradation.
Keywords: HCV; NS4B; PBM; Scribble; Tumorigenesis; Ubiquitin.
Conflict of interest statement
None
Figures



Similar articles
-
The ESEV PDZ-binding motif of the avian influenza A virus NS1 protein protects infected cells from apoptosis by directly targeting Scribble.J Virol. 2010 Nov;84(21):11164-74. doi: 10.1128/JVI.01278-10. Epub 2010 Aug 11. J Virol. 2010. PMID: 20702615 Free PMC article.
-
The avian influenza virus NS1 ESEV PDZ binding motif associates with Dlg1 and Scribble to disrupt cellular tight junctions.J Virol. 2011 Oct;85(20):10639-48. doi: 10.1128/JVI.05070-11. Epub 2011 Aug 17. J Virol. 2011. PMID: 21849460 Free PMC article.
-
Human T-cell leukemia virus type 1 Tax induces an aberrant clustering of the tumor suppressor Scribble through the PDZ domain-binding motif dependent and independent interaction.Virus Genes. 2008 Oct;37(2):231-40. doi: 10.1007/s11262-008-0259-4. Epub 2008 Jul 26. Virus Genes. 2008. PMID: 18661220
-
Glycine Zipper Motifs in Hepatitis C Virus Nonstructural Protein 4B Are Required for the Establishment of Viral Replication Organelles.J Virol. 2018 Jan 30;92(4):e01890-17. doi: 10.1128/JVI.01890-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29167346 Free PMC article.
-
HCV NS4B: From Obscurity to Central Stage.In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk (UK): Horizon Bioscience; 2006. Chapter 8. In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk (UK): Horizon Bioscience; 2006. Chapter 8. PMID: 21250395 Free Books & Documents. Review.
Cited by
-
The bZIP Proteins of Oncogenic Viruses.Viruses. 2020 Jul 14;12(7):757. doi: 10.3390/v12070757. Viruses. 2020. PMID: 32674309 Free PMC article. Review.
-
Hepatitis C virus NS4B protein induces epithelial-mesenchymal transition by upregulation of Snail.Virol J. 2017 Apr 21;14(1):83. doi: 10.1186/s12985-017-0737-1. Virol J. 2017. PMID: 28431572 Free PMC article.
-
Viral PDZ Binding Motifs Influence Cell Behavior Through the Interaction with Cellular Proteins Containing PDZ Domains.Methods Mol Biol. 2021;2256:217-236. doi: 10.1007/978-1-0716-1166-1_13. Methods Mol Biol. 2021. PMID: 34014525 Review.
-
Hepatitis C virus alters the morphology and function of peroxisomes.Front Microbiol. 2023 Sep 21;14:1254728. doi: 10.3389/fmicb.2023.1254728. eCollection 2023. Front Microbiol. 2023. PMID: 37808318 Free PMC article.
-
Viral subversion of the cell polarity regulator Scribble.Biochem Soc Trans. 2023 Feb 27;51(1):415-426. doi: 10.1042/BST20221067. Biochem Soc Trans. 2023. PMID: 36606695 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

