Splicing controls the ubiquitin response during DNA double-strand break repair
- PMID: 27315300
- PMCID: PMC5041194
- DOI: 10.1038/cdd.2016.58
Splicing controls the ubiquitin response during DNA double-strand break repair
Abstract
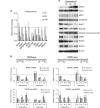
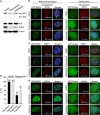
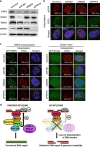
Although evidence that splicing regulates DNA repair is accumulating, the underlying mechanism(s) remain unclear. Here, we report that short-term inhibition of pre-mRNA splicing by spliceosomal inhibitors impairs cellular repair of DNA double-strand breaks. Indeed, interference with splicing as little as 1 h prior to irradiation reduced ubiquitylation of damaged chromatin and impaired recruitment of the repair factors WRAP53β, RNF168, 53BP1, BRCA1 and RAD51 to sites of DNA damage. Consequently, splicing-deficient cells exhibited significant numbers of residual γH2AX foci, as would be expected if DNA repair is defective. Furthermore, we show that this is due to downregulation of the E3 ubiquitin ligase RNF8 and that re-introduction of this protein into splicing-deficient cells restores ubiquitylation at sites of DNA damage, accumulation of downstream factors and subsequent repair. Moreover, downregulation of RNF8 explains the defective repair associated with knockdown of various splicing factors in recent genome-wide siRNA screens and, significantly, overexpression of RNF8 counteracts this defect. These discoveries reveal a mechanism that may not only explain how splicing regulates repair of double-strand breaks, but also may underlie various diseases caused by deregulation of splicing factors, including cancer.
Figures

Comment in
-
RNF8 - The Achilles heel of DNA repair when splicing rules.Cell Cycle. 2018;17(2):137-138. doi: 10.1080/15384101.2017.1360637. Epub 2018 Jan 18. Cell Cycle. 2018. PMID: 29323619 Free PMC article. No abstract available.
References
-
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131: 887–900. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

